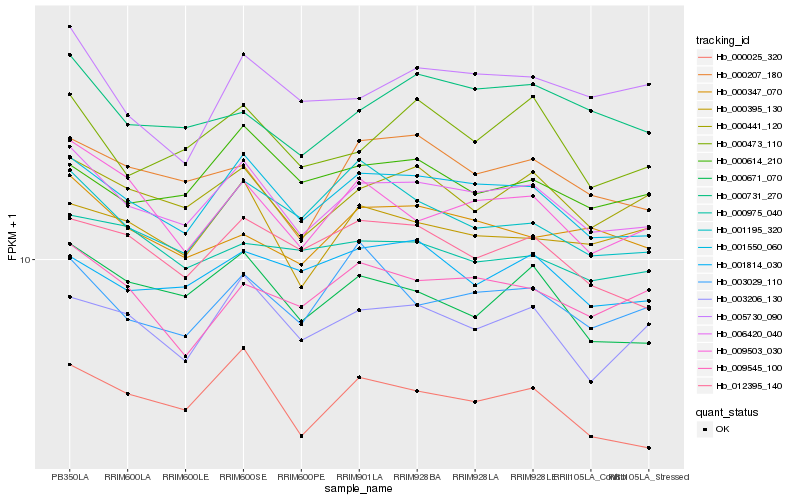
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006420_040 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001550_060 |
0.0279630462 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000441_120 |
0.0499278462 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000671_070 |
0.058309347 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 5 |
Hb_000025_320 |
0.058404606 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 6 |
Hb_003206_130 |
0.0599292286 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 7 |
Hb_009503_030 |
0.0611262254 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 8 |
Hb_000395_130 |
0.0614303268 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 9 |
Hb_012395_140 |
0.0616733814 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 10 |
Hb_000207_180 |
0.0620512099 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 11 |
Hb_000473_110 |
0.0623424403 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 12 |
Hb_000731_270 |
0.0642871564 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 13 |
Hb_000347_070 |
0.0645600031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001195_320 |
0.0666287733 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 15 |
Hb_000614_210 |
0.066656627 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_009545_100 |
0.0667670206 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 17 |
Hb_003029_110 |
0.0668709568 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001814_030 |
0.0671575318 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 19 |
Hb_005730_090 |
0.0689620326 |
- |
- |
PREDICTED: uncharacterized protein LOC105631484 [Jatropha curcas] |
| 20 |
Hb_000975_040 |
0.0692628499 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |