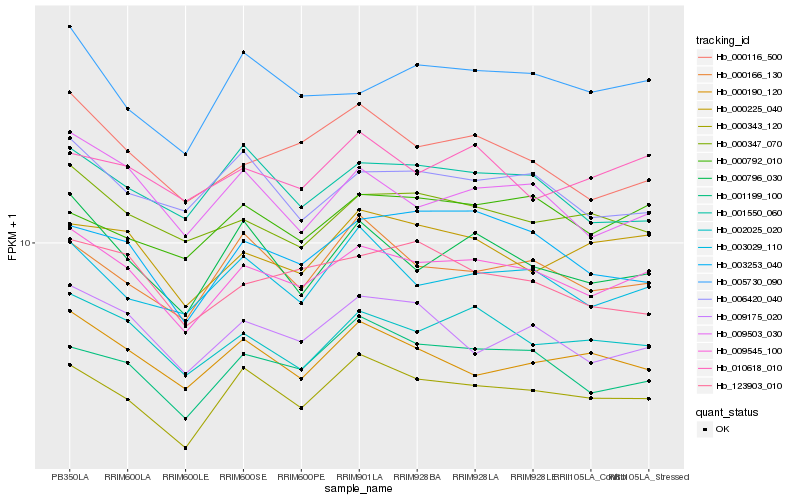
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009545_100 |
0.0 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 2 |
Hb_002025_020 |
0.0512228789 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 3 |
Hb_000343_120 |
0.0554212362 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 4 |
Hb_000347_070 |
0.0594194302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000116_500 |
0.0621292288 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 6 |
Hb_003029_110 |
0.0624752371 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000225_040 |
0.0624839617 |
- |
- |
PREDICTED: translation factor GUF1 homolog, mitochondrial [Nelumbo nucifera] |
| 8 |
Hb_009175_020 |
0.0632314736 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_001199_100 |
0.063271721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 10 |
Hb_009503_030 |
0.0639499733 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 11 |
Hb_005730_090 |
0.0642268768 |
- |
- |
PREDICTED: uncharacterized protein LOC105631484 [Jatropha curcas] |
| 12 |
Hb_000796_030 |
0.0658755074 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_006420_040 |
0.0667670206 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000190_120 |
0.0669475406 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 15 |
Hb_001550_060 |
0.0669901157 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 16 |
Hb_000166_130 |
0.0680388602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010618_010 |
0.0687188216 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 18 |
Hb_003253_040 |
0.0690502846 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 19 |
Hb_000792_010 |
0.0694740816 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 20 |
Hb_123903_010 |
0.0699368279 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |