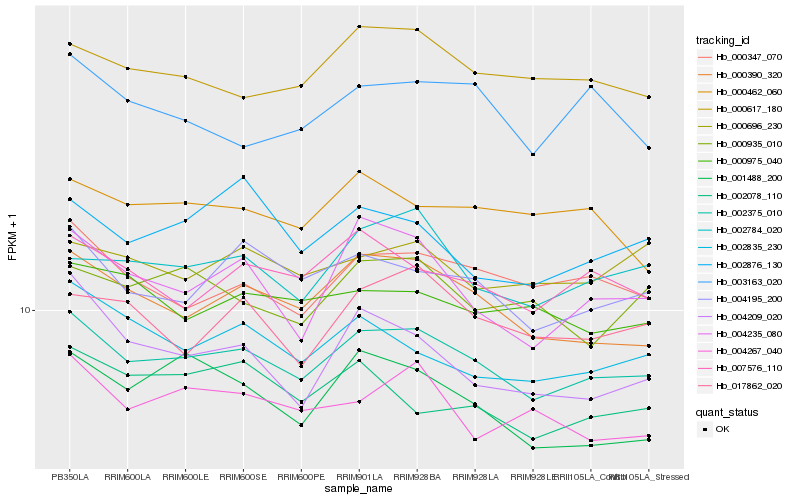
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002375_010 |
0.0 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000390_320 |
0.0472735646 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 3 |
Hb_004195_200 |
0.0521242514 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 4 |
Hb_000347_070 |
0.0555551501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007576_110 |
0.0573617744 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 6 |
Hb_000617_180 |
0.0592116834 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 7 |
Hb_002784_020 |
0.0621014419 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_004235_080 |
0.0637523075 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 9 |
Hb_002835_230 |
0.0651961839 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000975_040 |
0.0655859118 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 11 |
Hb_017862_020 |
0.0662381976 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 12 |
Hb_000696_230 |
0.0669007003 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 13 |
Hb_000935_010 |
0.0671803905 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 14 |
Hb_000462_060 |
0.067880111 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004267_040 |
0.0689827745 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 16 |
Hb_003163_020 |
0.0693035996 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002078_110 |
0.0696212145 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 18 |
Hb_002876_130 |
0.070389727 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 19 |
Hb_001488_200 |
0.0707681336 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_004209_020 |
0.0716599239 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |