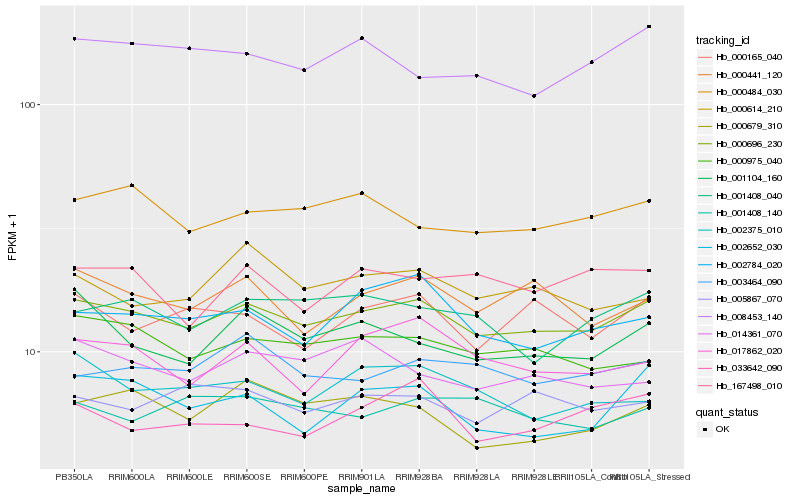
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_230 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 2 |
Hb_000975_040 |
0.0582570752 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 3 |
Hb_000484_030 |
0.0602396639 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 4 |
Hb_000441_120 |
0.0621129405 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001408_040 |
0.0631759312 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005867_070 |
0.0634699932 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_003464_090 |
0.0637108028 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 8 |
Hb_000679_310 |
0.0642412438 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002784_020 |
0.0645617045 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_008453_140 |
0.066225527 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 11 |
Hb_001408_140 |
0.0663339794 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 12 |
Hb_000165_040 |
0.0666811839 |
- |
- |
hypothetical protein JCGZ_01206 [Jatropha curcas] |
| 13 |
Hb_002375_010 |
0.0669007003 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 14 |
Hb_033642_090 |
0.0670008606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002652_030 |
0.0674373946 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_017862_020 |
0.0681726354 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 17 |
Hb_000614_210 |
0.0687100033 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001104_160 |
0.0688263537 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 19 |
Hb_014361_070 |
0.0692235104 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 20 |
Hb_167498_010 |
0.0693060287 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |