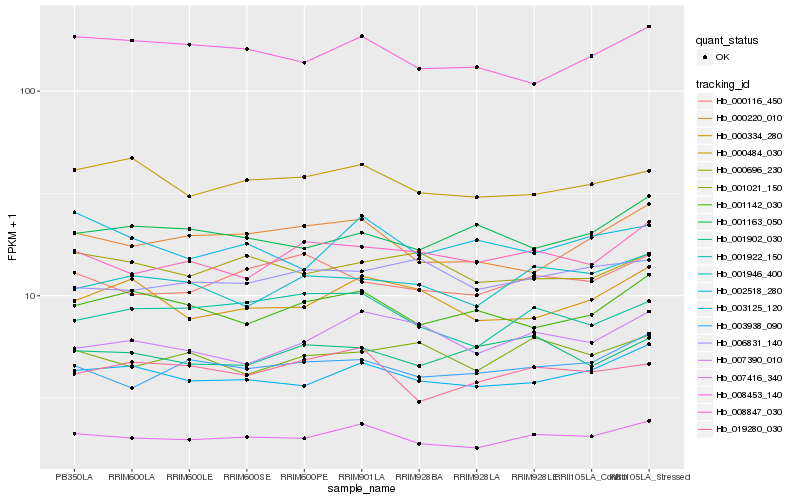
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_003125_120 |
0.0428949752 |
- |
- |
PREDICTED: TITAN-like protein [Jatropha curcas] |
| 3 |
Hb_003938_090 |
0.0568126294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000484_030 |
0.0601854568 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 5 |
Hb_008453_140 |
0.0615255422 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 6 |
Hb_001946_400 |
0.0618927926 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001922_150 |
0.0632696621 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_019280_030 |
0.0632981276 |
- |
- |
- |
| 9 |
Hb_000220_010 |
0.0636376033 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 10 |
Hb_008847_030 |
0.0647006807 |
- |
- |
PREDICTED: TLD domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_001142_030 |
0.0670750083 |
- |
- |
PREDICTED: uncharacterized protein LOC105640023 [Jatropha curcas] |
| 12 |
Hb_000116_450 |
0.0685519713 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_007390_010 |
0.0687373065 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000334_280 |
0.0696851961 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 15 |
Hb_000696_230 |
0.0700548444 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 16 |
Hb_001902_030 |
0.0706397935 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001021_150 |
0.0706608979 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 18 |
Hb_006831_140 |
0.0716343889 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_002518_280 |
0.0720266325 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 20 |
Hb_001163_050 |
0.0720604878 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |