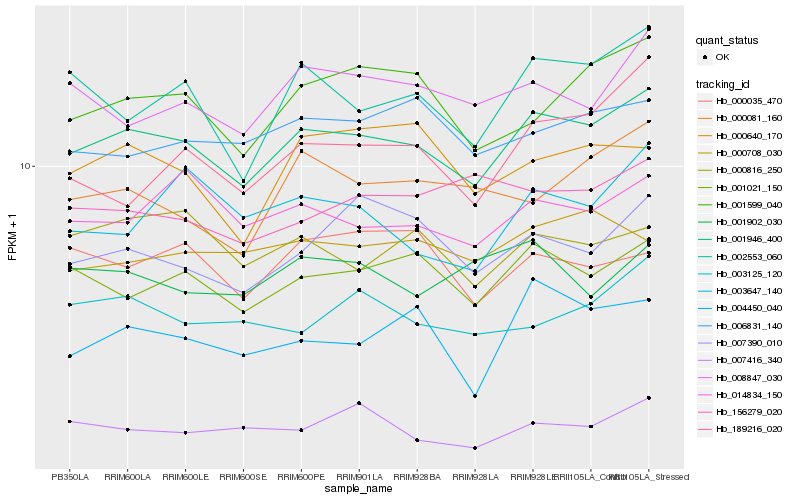
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_400 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001021_150 |
0.0512682403 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 3 |
Hb_014834_150 |
0.0597070401 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001599_040 |
0.0602449736 |
- |
- |
PREDICTED: suppressor protein SRP40 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002553_060 |
0.0602665147 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit beta, mitochondrial [Jatropha curcas] |
| 6 |
Hb_008847_030 |
0.0614923856 |
- |
- |
PREDICTED: TLD domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_007416_340 |
0.0618927926 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_006831_140 |
0.065029726 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004450_040 |
0.0662633817 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 10 |
Hb_000816_250 |
0.0665469778 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007390_010 |
0.0667516978 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_003125_120 |
0.0672303737 |
- |
- |
PREDICTED: TITAN-like protein [Jatropha curcas] |
| 13 |
Hb_000708_030 |
0.0682626934 |
- |
- |
PREDICTED: uncharacterized protein LOC105650742 [Jatropha curcas] |
| 14 |
Hb_000081_160 |
0.0700378303 |
- |
- |
PREDICTED: uncharacterized protein At4g17910 [Jatropha curcas] |
| 15 |
Hb_000640_170 |
0.0707211185 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000035_470 |
0.0715132612 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001902_030 |
0.072017024 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003647_140 |
0.0725437993 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 19 |
Hb_189216_020 |
0.0728739856 |
- |
- |
PREDICTED: translin [Jatropha curcas] |
| 20 |
Hb_156279_020 |
0.0731678971 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |