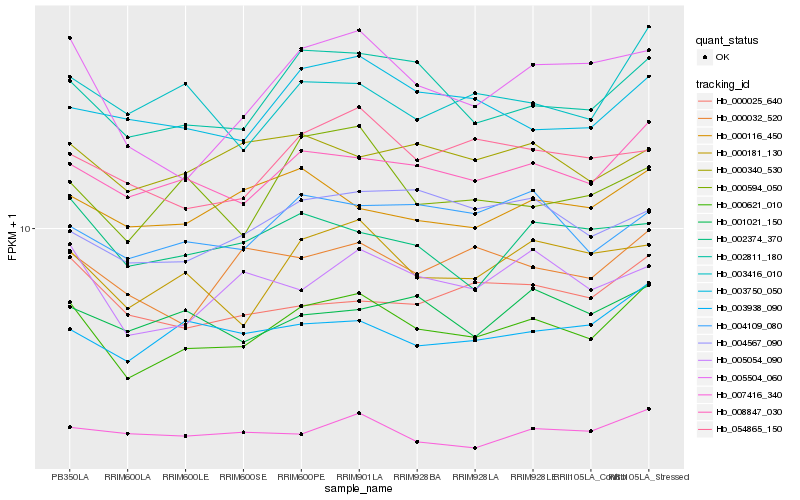
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000621_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103444129 [Malus domestica] |
| 2 |
Hb_008847_030 |
0.0563298259 |
- |
- |
PREDICTED: TLD domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_002811_180 |
0.0616372956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000181_130 |
0.0702777264 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003938_090 |
0.0746791249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005054_090 |
0.0761074484 |
- |
- |
PREDICTED: uncharacterized protein At2g39910 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005504_060 |
0.0767665757 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Populus euphratica] |
| 8 |
Hb_003750_050 |
0.0801142398 |
- |
- |
PREDICTED: THO complex subunit 4D isoform X1 [Jatropha curcas] |
| 9 |
Hb_000032_520 |
0.0815279996 |
- |
- |
PREDICTED: F-box protein SKIP8-like [Jatropha curcas] |
| 10 |
Hb_007416_340 |
0.0819022544 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_004109_080 |
0.0820191841 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 12 |
Hb_003416_010 |
0.0822904199 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 13 homolog B [Malus domestica] |
| 13 |
Hb_054865_150 |
0.0825861066 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein Q [Jatropha curcas] |
| 14 |
Hb_000116_450 |
0.0831286121 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_000025_640 |
0.0832626371 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002374_370 |
0.0842151428 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_004567_090 |
0.0847517389 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 18 |
Hb_000594_050 |
0.0848539642 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001021_150 |
0.086142535 |
- |
- |
PREDICTED: metal tolerance protein C1 [Jatropha curcas] |
| 20 |
Hb_000340_530 |
0.0871529583 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |