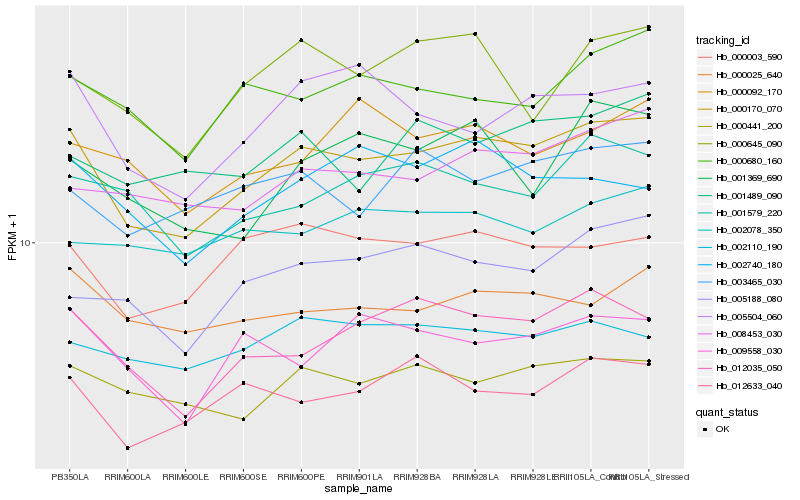
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_070 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 2 |
Hb_000441_200 |
0.0812916306 |
- |
- |
PREDICTED: small RNA 2'-O-methyltransferase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_012035_050 |
0.0828012401 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 4 |
Hb_000003_590 |
0.0860431187 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000025_640 |
0.0861442163 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005504_060 |
0.0882526816 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Populus euphratica] |
| 7 |
Hb_008453_030 |
0.0883791645 |
- |
- |
Thioredoxin superfamily protein [Theobroma cacao] |
| 8 |
Hb_005188_080 |
0.0911193464 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 9 |
Hb_001579_220 |
0.0913121996 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 10 |
Hb_000680_160 |
0.0918254431 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 11 |
Hb_003465_030 |
0.0922135655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000645_090 |
0.0936356377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002110_190 |
0.0944923487 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 14 |
Hb_012633_040 |
0.0953320378 |
- |
- |
- |
| 15 |
Hb_001489_090 |
0.0958619128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649775 [Jatropha curcas] |
| 16 |
Hb_009558_030 |
0.0977226703 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 17 |
Hb_001369_690 |
0.0984038223 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 18 |
Hb_002740_180 |
0.0988168232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000092_170 |
0.0993938575 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_002078_350 |
0.0995702399 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |