| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
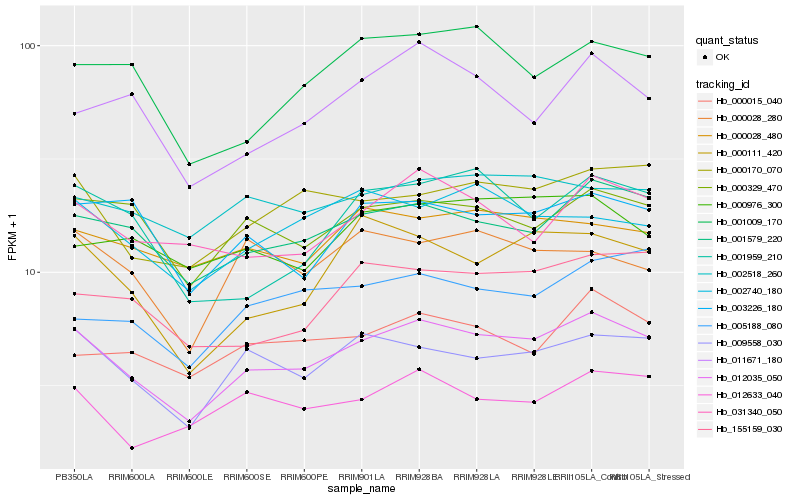
Hb_012035_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 2 |
Hb_001579_220 |
0.0737098679 |
- |
- |
PREDICTED: pantothenate kinase 1 [Jatropha curcas] |
| 3 |
Hb_009558_030 |
0.0767205412 |
- |
- |
myb3r3, putative [Ricinus communis] |
| 4 |
Hb_000170_070 |
0.0828012401 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 5 |
Hb_000111_420 |
0.0891860915 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 6 |
Hb_000329_470 |
0.0899450846 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 7 |
Hb_155159_030 |
0.09323379 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 8 |
Hb_031340_050 |
0.0936724269 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001009_170 |
0.0962523489 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_003226_180 |
0.0963956253 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 11 |
Hb_002740_180 |
0.097160382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001959_210 |
0.097840204 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 13 |
Hb_012633_040 |
0.0990968229 |
- |
- |
- |
| 14 |
Hb_000028_480 |
0.1007866501 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 15 |
Hb_002518_260 |
0.1009010874 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 16 |
Hb_000015_040 |
0.1009269816 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011671_180 |
0.1014013418 |
- |
- |
PREDICTED: probable quinone oxidoreductase [Jatropha curcas] |
| 18 |
Hb_000028_280 |
0.1029724809 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 19 |
Hb_000976_300 |
0.1031027644 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 20 |
Hb_005188_080 |
0.1031429228 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |