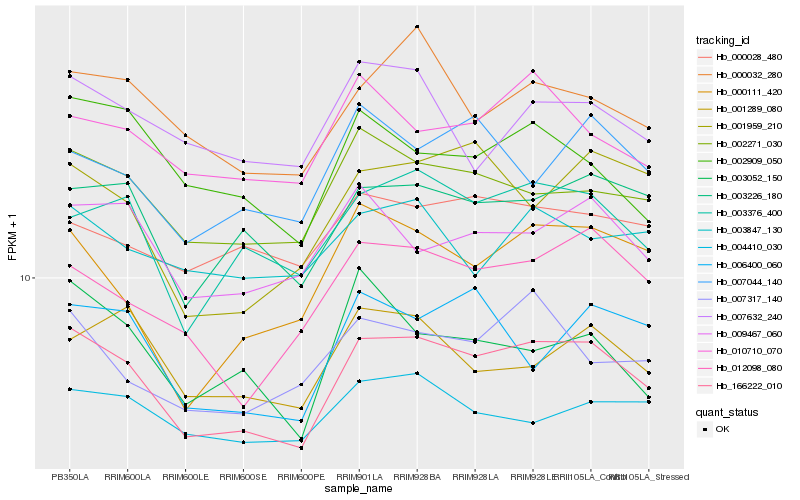
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166222_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 2 |
Hb_002271_030 |
0.0830028995 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 3 |
Hb_003226_180 |
0.0830387154 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 4 |
Hb_003376_400 |
0.0840452185 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 5 |
Hb_012098_080 |
0.0879063601 |
- |
- |
KOM, putative [Ricinus communis] |
| 6 |
Hb_000032_280 |
0.0924056279 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 7 |
Hb_000111_420 |
0.0955360084 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 8 |
Hb_007317_140 |
0.0963436636 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 9 |
Hb_009467_060 |
0.0964617732 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001959_210 |
0.0978313775 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 11 |
Hb_006400_060 |
0.099797581 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010710_070 |
0.1009145764 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_001289_080 |
0.1020970757 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 14 |
Hb_002909_050 |
0.1023839036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004410_030 |
0.1030805294 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_007044_140 |
0.104157019 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta [Jatropha curcas] |
| 17 |
Hb_007632_240 |
0.1049962659 |
- |
- |
transporter, putative [Ricinus communis] |
| 18 |
Hb_000028_480 |
0.1053119333 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 19 |
Hb_003052_150 |
0.1060133578 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 20 |
Hb_003847_130 |
0.1065333873 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |