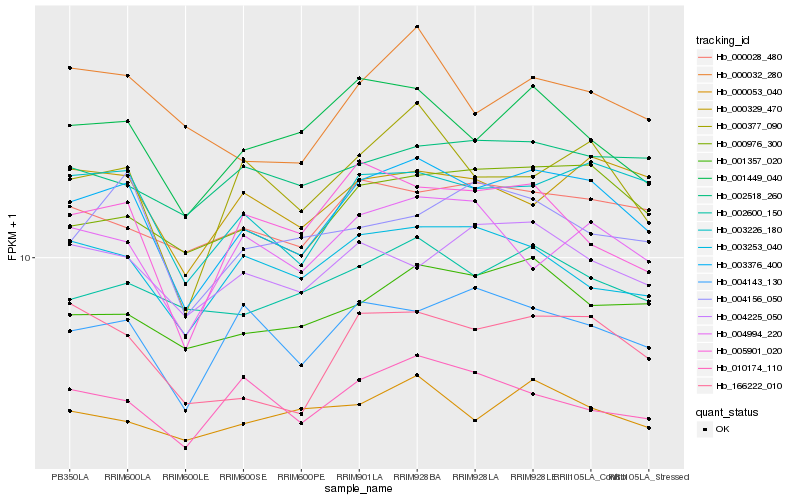
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_400 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 2 |
Hb_003226_180 |
0.0714409526 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 3 |
Hb_000976_300 |
0.0799263203 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 4 |
Hb_166222_010 |
0.0840452185 |
- |
- |
hypothetical protein POPTR_0017s08350g [Populus trichocarpa] |
| 5 |
Hb_004143_130 |
0.0857847545 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |
| 6 |
Hb_000377_090 |
0.0858271483 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 7 |
Hb_001449_040 |
0.0869647382 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001357_020 |
0.0895321232 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002600_150 |
0.0911974644 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000028_480 |
0.0919425505 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 11 |
Hb_003253_040 |
0.0926267643 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 12 |
Hb_004225_050 |
0.0932944217 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000329_470 |
0.093504556 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 14 |
Hb_004994_220 |
0.0941740446 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_005901_020 |
0.0961000838 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_010174_110 |
0.097902276 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 17 |
Hb_000053_040 |
0.0989344062 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 18 |
Hb_004156_050 |
0.1008102664 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 19 |
Hb_002518_260 |
0.1010866113 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 20 |
Hb_000032_280 |
0.1016257511 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |