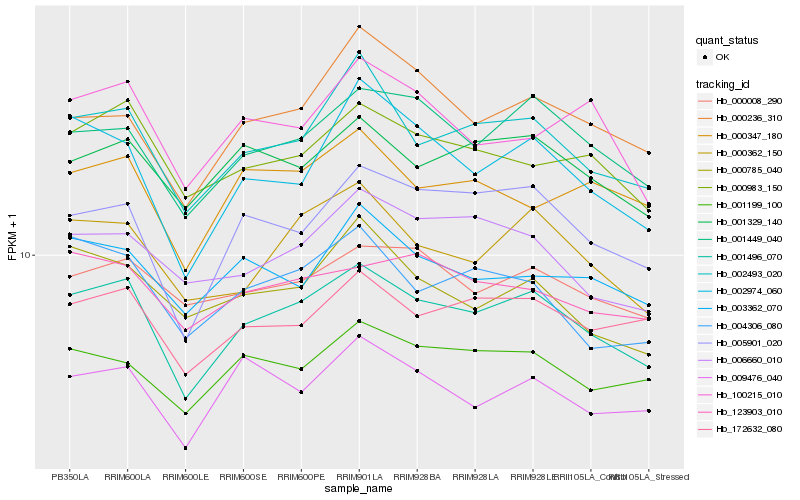
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001496_070 |
0.0 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 2 |
Hb_002493_020 |
0.069227036 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 3 |
Hb_005901_020 |
0.0706169825 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_001449_040 |
0.0715621907 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000362_150 |
0.0753261979 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 6 |
Hb_000983_150 |
0.0813057432 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 7 |
Hb_002974_060 |
0.0815114183 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 8 |
Hb_172632_080 |
0.0836418606 |
- |
- |
PREDICTED: uncharacterized protein LOC105141138 isoform X1 [Populus euphratica] |
| 9 |
Hb_001329_140 |
0.0858014312 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 10 |
Hb_001199_100 |
0.0863993383 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 11 |
Hb_004306_080 |
0.0887539706 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 12 |
Hb_006660_010 |
0.0922146983 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 13 |
Hb_009476_040 |
0.0924363942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000347_180 |
0.0927311528 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 15 |
Hb_000008_290 |
0.0928569339 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 16 |
Hb_123903_010 |
0.0929257156 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000236_310 |
0.0935132288 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 18 |
Hb_003362_070 |
0.0939387184 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 19 |
Hb_100215_010 |
0.0941089728 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 20 |
Hb_000785_040 |
0.0951555842 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |