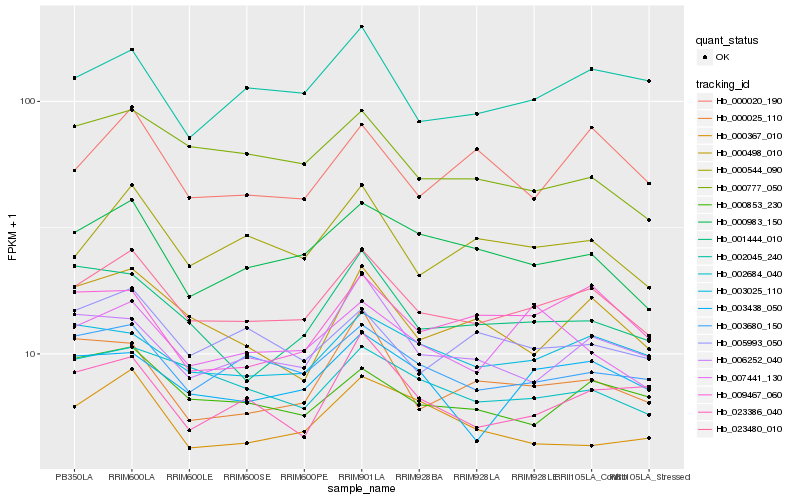
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023480_010 |
0.0 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_003680_150 |
0.0621866421 |
- |
- |
srpk, putative [Ricinus communis] |
| 3 |
Hb_000983_150 |
0.0642260854 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 4 |
Hb_000544_090 |
0.0645691133 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 5 |
Hb_002684_040 |
0.066709949 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003025_110 |
0.0678225489 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000853_230 |
0.0703377536 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_002045_240 |
0.0710014534 |
- |
- |
2-methyl-6-phytylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 9 |
Hb_003438_050 |
0.0723373036 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 10 |
Hb_007441_130 |
0.0736177421 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_006252_040 |
0.0736420649 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005993_050 |
0.0741166781 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 13 |
Hb_000498_010 |
0.074252668 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000367_010 |
0.0752371894 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 15 |
Hb_000025_110 |
0.0762644247 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 16 |
Hb_000777_050 |
0.0768848829 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 17 |
Hb_009467_060 |
0.0774421229 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_023386_040 |
0.0779147548 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 19 |
Hb_001444_010 |
0.0787839705 |
transcription factor |
TF Family: DDT |
PREDICTED: DDT domain-containing protein DDB_G0282237 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000020_190 |
0.0788016219 |
- |
- |
hypothetical protein RCOM_0452240 [Ricinus communis] |