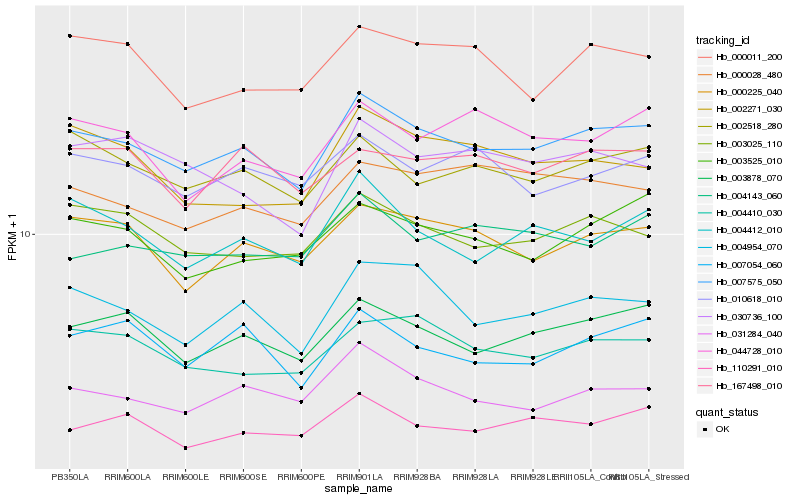
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007575_050 |
0.0 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 2 |
Hb_003878_070 |
0.0509762098 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 3 |
Hb_000225_040 |
0.0577514736 |
- |
- |
PREDICTED: translation factor GUF1 homolog, mitochondrial [Nelumbo nucifera] |
| 4 |
Hb_004954_070 |
0.0580918814 |
- |
- |
PREDICTED: uncharacterized protein LOC105645187 [Jatropha curcas] |
| 5 |
Hb_003025_110 |
0.0584439466 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002518_280 |
0.060436495 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_004412_010 |
0.0623856732 |
- |
- |
- |
| 8 |
Hb_004410_030 |
0.0643839295 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_031284_040 |
0.0647584426 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 10 |
Hb_003525_010 |
0.0653088466 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 24-like isoform X1 [Populus euphratica] |
| 11 |
Hb_010618_010 |
0.0679357422 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 12 |
Hb_000011_200 |
0.0687689669 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_167498_010 |
0.0691111616 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 14 |
Hb_000028_480 |
0.0697039569 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 15 |
Hb_004143_060 |
0.0705367573 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_007054_060 |
0.0711910956 |
- |
- |
- |
| 17 |
Hb_030736_100 |
0.0715570295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_044728_010 |
0.0727105693 |
- |
- |
- |
| 19 |
Hb_002271_030 |
0.0747359237 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 20 |
Hb_110291_010 |
0.0753968237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |