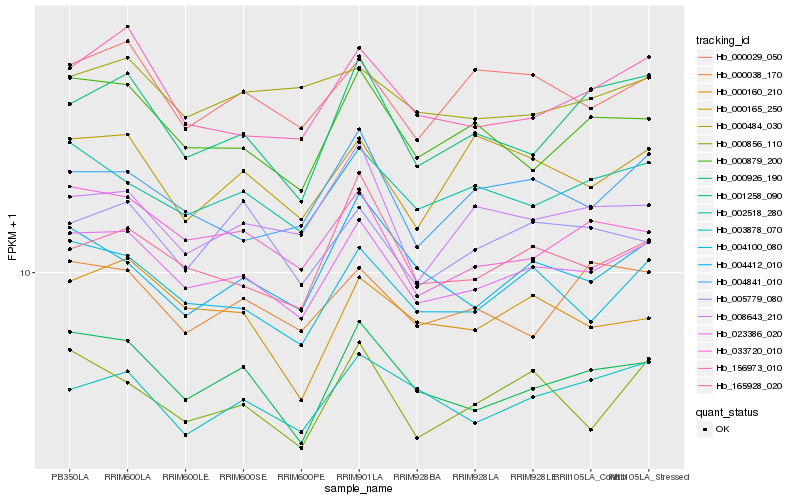
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023386_020 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004100_080 |
0.0437132771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000926_190 |
0.0498312096 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_002518_280 |
0.0520313747 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 5 |
Hb_033720_010 |
0.0528064256 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 6 |
Hb_156973_010 |
0.0555614524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000879_200 |
0.0563194857 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 8 |
Hb_000029_050 |
0.0587841187 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 9 |
Hb_004841_010 |
0.0599078099 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_165928_020 |
0.0607671351 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001258_090 |
0.0637813098 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 12 |
Hb_003878_070 |
0.0655119725 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 13 |
Hb_000856_110 |
0.065794808 |
- |
- |
PREDICTED: uncharacterized protein LOC105640477 isoform X2 [Jatropha curcas] |
| 14 |
Hb_008643_210 |
0.0663464384 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004412_010 |
0.0665723298 |
- |
- |
- |
| 16 |
Hb_000038_170 |
0.0671244384 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 17 |
Hb_000160_210 |
0.0674829143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005779_080 |
0.0694564094 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000484_030 |
0.0698168768 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 20 |
Hb_000165_250 |
0.0699900548 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |