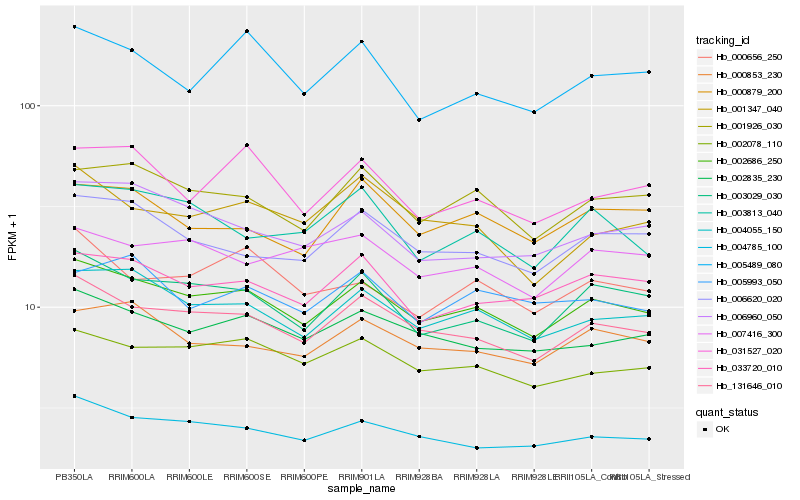
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_250 |
0.0 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 2 |
Hb_131646_010 |
0.0356635007 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_004055_150 |
0.0418710954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003029_030 |
0.051793541 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004785_100 |
0.0520389229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001926_030 |
0.0529404509 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 7 |
Hb_002078_110 |
0.0537981453 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 8 |
Hb_002835_230 |
0.0541143309 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_033720_010 |
0.0593992162 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 10 |
Hb_000853_230 |
0.0598501277 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_006620_020 |
0.06011851 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 12 |
Hb_006960_050 |
0.0665902943 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 13 |
Hb_000879_200 |
0.0697815949 |
- |
- |
PREDICTED: A-kinase anchor protein 17A [Jatropha curcas] |
| 14 |
Hb_031527_020 |
0.0718131992 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 15 |
Hb_003813_040 |
0.0721729438 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 16 |
Hb_000656_250 |
0.0732113107 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 17 |
Hb_005489_080 |
0.0736377494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007416_300 |
0.0737885996 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 19 |
Hb_005993_050 |
0.0739440136 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 20 |
Hb_001347_040 |
0.0746010727 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |