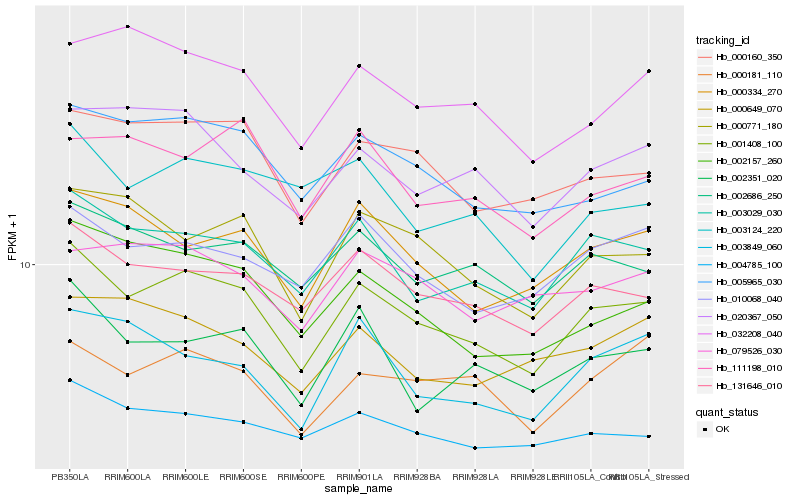
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_100 |
0.0 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 2 |
Hb_003029_030 |
0.0644518446 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 3 |
Hb_131646_010 |
0.0686641838 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000160_350 |
0.0756659918 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 5 |
Hb_005965_030 |
0.0781143134 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 6 |
Hb_002157_260 |
0.080139632 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 7 |
Hb_000771_180 |
0.0810199765 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 8 |
Hb_004785_100 |
0.0821299306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000334_270 |
0.0831943509 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_010068_040 |
0.0835368022 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 11 |
Hb_002686_250 |
0.0837170445 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 12 |
Hb_002351_020 |
0.0869824823 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 13 |
Hb_020367_050 |
0.0871566959 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_003124_220 |
0.0910770769 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 15 |
Hb_032208_040 |
0.0911453951 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 16 |
Hb_000181_110 |
0.0911829775 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 17 |
Hb_003849_060 |
0.0943892073 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 18 |
Hb_079526_030 |
0.0953711742 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 19 |
Hb_000649_070 |
0.0960253779 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 20 |
Hb_111198_010 |
0.0966710147 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |