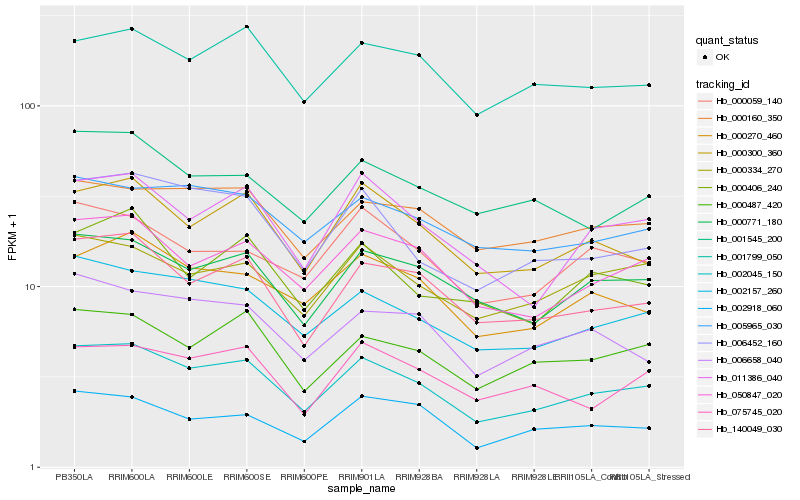
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_150 |
0.0 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 2 |
Hb_002157_260 |
0.0607222558 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 3 |
Hb_140049_030 |
0.0637727504 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 4 |
Hb_050847_020 |
0.0660723688 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_000771_180 |
0.0704799302 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 6 |
Hb_000300_360 |
0.0711469805 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 7 |
Hb_006452_160 |
0.071633372 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 8 |
Hb_011386_040 |
0.0734377631 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000487_420 |
0.0752223836 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 10 |
Hb_000270_460 |
0.0753671301 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 11 |
Hb_000334_270 |
0.0818080069 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005965_030 |
0.0832571153 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 13 |
Hb_000160_350 |
0.0842733315 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 14 |
Hb_001799_050 |
0.084953854 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 15 |
Hb_006658_040 |
0.0852206757 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 16 |
Hb_000059_140 |
0.0865184483 |
- |
- |
rnase h, putative [Ricinus communis] |
| 17 |
Hb_075745_020 |
0.0870573212 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002918_060 |
0.0873896547 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_001545_200 |
0.0874986129 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 20 |
Hb_000406_240 |
0.0875367355 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |