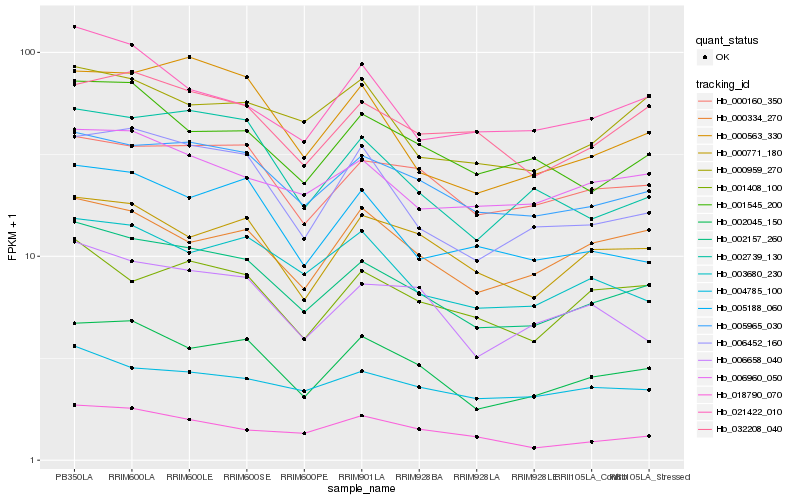
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_260 |
0.0 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 2 |
Hb_005965_030 |
0.0534358683 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 3 |
Hb_002045_150 |
0.0607222558 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 4 |
Hb_000160_350 |
0.0733302645 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 5 |
Hb_006452_160 |
0.0775353577 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 6 |
Hb_006960_050 |
0.0788042366 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_001408_100 |
0.080139632 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 8 |
Hb_001545_200 |
0.0801473456 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 9 |
Hb_000771_180 |
0.0814052127 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 10 |
Hb_004785_100 |
0.0815491497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_032208_040 |
0.0816345344 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 12 |
Hb_003680_230 |
0.0818437407 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 13 |
Hb_006658_040 |
0.0818447421 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |
| 14 |
Hb_005188_060 |
0.0836325375 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 15 |
Hb_002739_130 |
0.0841265158 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 16 |
Hb_021422_010 |
0.0847654749 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000563_330 |
0.085943683 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000959_270 |
0.0866725008 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 19 |
Hb_018790_070 |
0.0868730637 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 20 |
Hb_000334_270 |
0.0889521697 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |