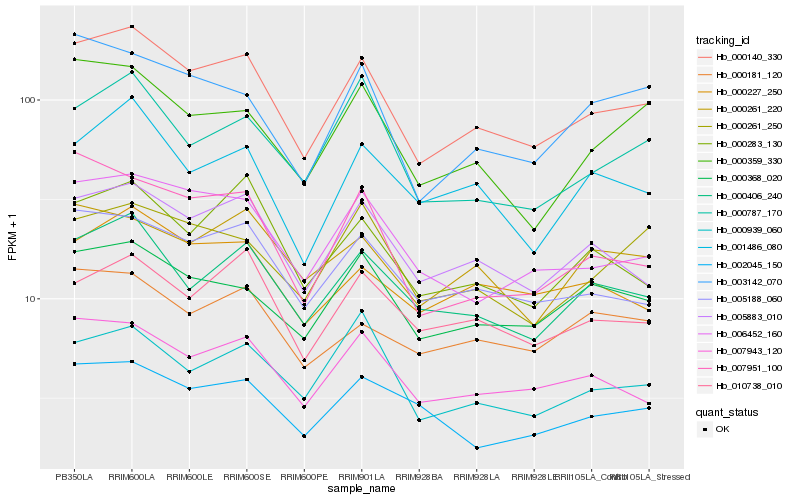
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_330 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 2 |
Hb_005883_010 |
0.0668166041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005188_060 |
0.0702311308 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 4 |
Hb_007943_120 |
0.0728714097 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000406_240 |
0.0738574557 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 6 |
Hb_006452_160 |
0.0753212623 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 7 |
Hb_000368_020 |
0.0814118341 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_010738_010 |
0.0872176356 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 9 |
Hb_000283_130 |
0.0875626218 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 10 |
Hb_000227_250 |
0.0888504603 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 11 |
Hb_000359_330 |
0.0904887494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001486_080 |
0.0920914176 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 13 |
Hb_003142_070 |
0.092404171 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 14 |
Hb_007951_100 |
0.0927036534 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000787_170 |
0.0928328932 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 16 |
Hb_000939_060 |
0.0938848102 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 17 |
Hb_000261_220 |
0.0945860612 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 18 |
Hb_000261_250 |
0.0946578446 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 19 |
Hb_000181_120 |
0.0946781641 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 20 |
Hb_002045_150 |
0.0950404478 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |