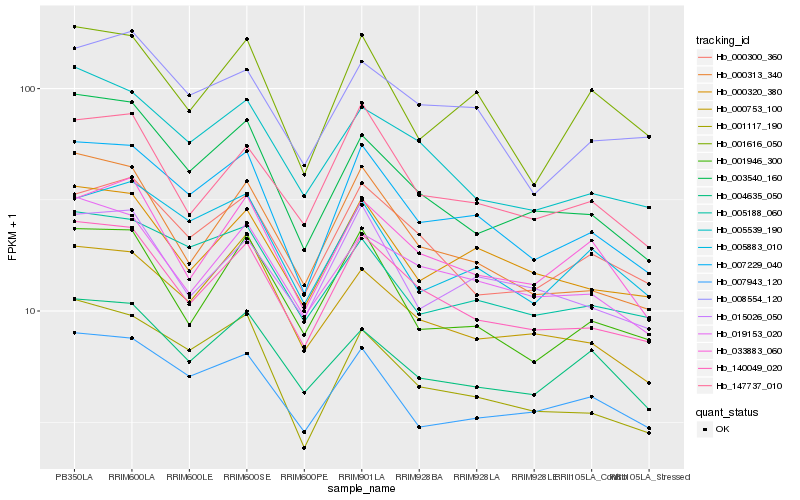
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007229_040 |
0.0 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 2 |
Hb_140049_020 |
0.0606442662 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 3 |
Hb_001117_190 |
0.0616733438 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 4 |
Hb_005883_010 |
0.0698241924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000320_380 |
0.0708658132 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005188_060 |
0.072275651 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_007943_120 |
0.0723921902 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 8 |
Hb_001616_050 |
0.073537541 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 9 |
Hb_008554_120 |
0.0781241567 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 10 |
Hb_000753_100 |
0.0787023619 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000300_360 |
0.07880471 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 12 |
Hb_003540_160 |
0.0803227294 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 13 |
Hb_000313_340 |
0.0817855861 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 14 |
Hb_019153_020 |
0.0818839581 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001946_300 |
0.0840457911 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 16 |
Hb_005539_190 |
0.0840594332 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 17 |
Hb_015026_050 |
0.0845851423 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 18 |
Hb_033883_060 |
0.085196512 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 19 |
Hb_147737_010 |
0.0852664189 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004635_050 |
0.0856312671 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |