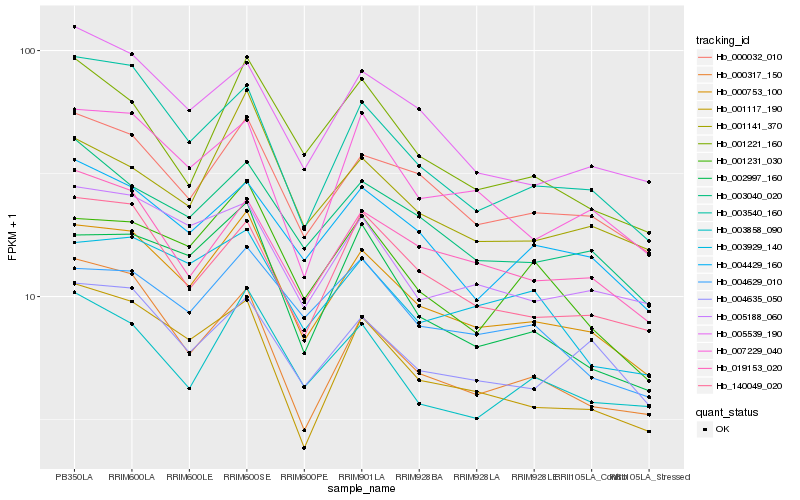
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_100 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000032_010 |
0.0582154552 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 3 |
Hb_003540_160 |
0.0665667369 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 4 |
Hb_003040_020 |
0.0713858984 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 5 |
Hb_001117_190 |
0.0736294854 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 6 |
Hb_140049_020 |
0.0747632219 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 7 |
Hb_005188_060 |
0.0768688438 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 8 |
Hb_019153_020 |
0.0768706704 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004429_160 |
0.0769386927 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005539_190 |
0.0778058427 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 11 |
Hb_001231_030 |
0.0781577691 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 12 |
Hb_002997_160 |
0.0785023025 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007229_040 |
0.0787023619 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 14 |
Hb_004635_050 |
0.0794190685 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_003929_140 |
0.079625312 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_003858_090 |
0.0802393622 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 17 |
Hb_004629_010 |
0.0813341082 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 18 |
Hb_001141_370 |
0.0816748063 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 19 |
Hb_000317_150 |
0.0850569836 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_001221_160 |
0.0852749865 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |