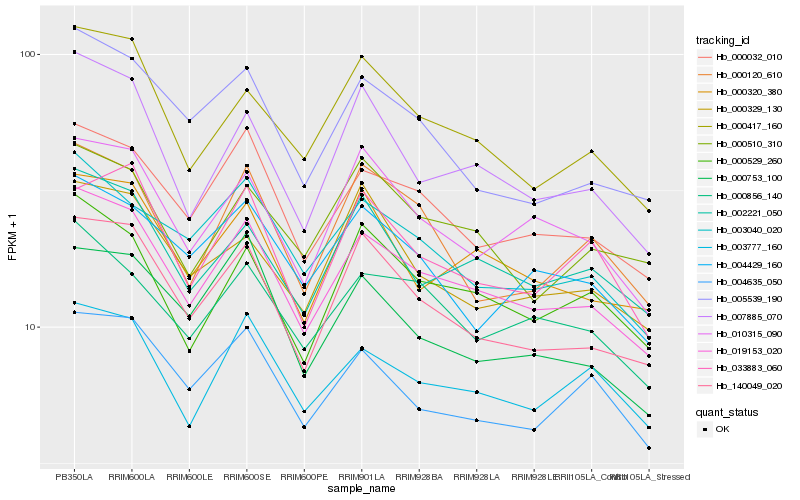
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019153_020 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000032_010 |
0.0488591725 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 3 |
Hb_140049_020 |
0.0538930963 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 4 |
Hb_000417_160 |
0.0603878763 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 5 |
Hb_002221_050 |
0.0606863343 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000529_260 |
0.0619441021 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 7 |
Hb_010315_090 |
0.0622866815 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 8 |
Hb_000856_140 |
0.0627546545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003777_160 |
0.0630724856 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000320_380 |
0.0633240794 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007885_070 |
0.0634199841 |
- |
- |
cleavage and polyadenylation specificity factor, putative [Ricinus communis] |
| 12 |
Hb_003040_020 |
0.0662426303 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 13 |
Hb_005539_190 |
0.0707121353 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 14 |
Hb_004635_050 |
0.0708872353 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_004429_160 |
0.0756913763 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000510_310 |
0.075864649 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 17 |
Hb_033883_060 |
0.0761773214 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 18 |
Hb_000120_610 |
0.0765831545 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 19 |
Hb_000329_130 |
0.0767801601 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 20 |
Hb_000753_100 |
0.0768706704 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |