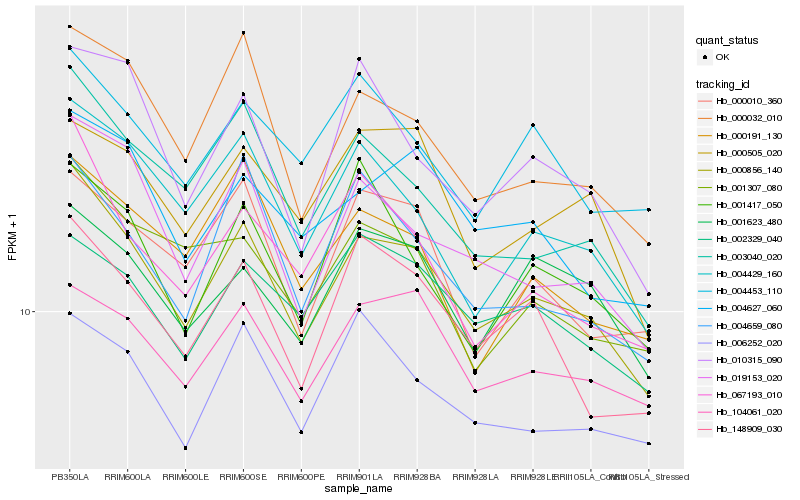
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000856_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000032_010 |
0.0622955796 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 3 |
Hb_019153_020 |
0.0627546545 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003040_020 |
0.066585521 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 5 |
Hb_004429_160 |
0.0672710982 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_148909_030 |
0.0702297936 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004627_060 |
0.0722510206 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 8 |
Hb_104061_020 |
0.0724218218 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 9 |
Hb_067193_010 |
0.0728137902 |
- |
- |
- |
| 10 |
Hb_004659_080 |
0.0749340398 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 11 |
Hb_010315_090 |
0.0759527086 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 12 |
Hb_000505_020 |
0.0761656288 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 13 |
Hb_006252_020 |
0.0769038989 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001623_480 |
0.0770060836 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 15 |
Hb_000010_360 |
0.0773062146 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 16 |
Hb_001307_080 |
0.0776922227 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 17 |
Hb_002329_040 |
0.0777217351 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 18 |
Hb_004453_110 |
0.0786508439 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000191_130 |
0.0794170385 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 20 |
Hb_001417_050 |
0.0799505563 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |