| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
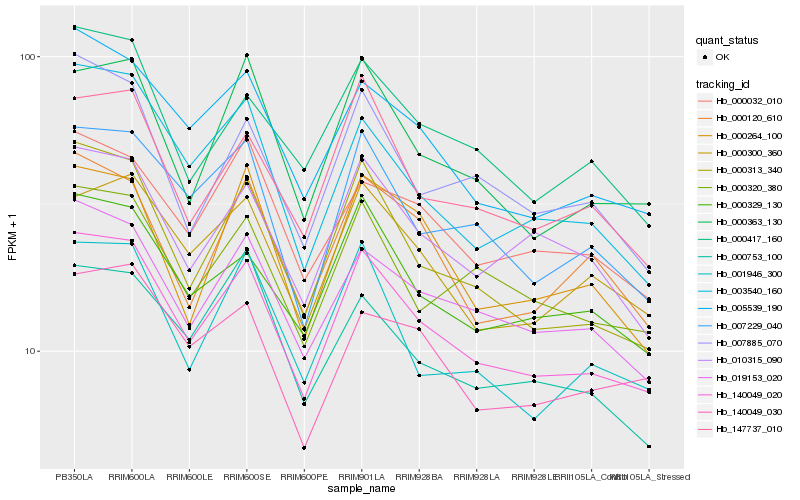
Hb_140049_020 |
0.0 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 2 |
Hb_000313_340 |
0.0505275018 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |
| 3 |
Hb_019153_020 |
0.0538930963 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005539_190 |
0.0542513949 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 5 |
Hb_000329_130 |
0.0584945852 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 6 |
Hb_007229_040 |
0.0606442662 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 7 |
Hb_000363_130 |
0.0624895828 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000300_360 |
0.0633818151 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 9 |
Hb_147737_010 |
0.0636725225 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003540_160 |
0.0647802539 |
- |
- |
hypothetical protein POPTR_0011s12160g [Populus trichocarpa] |
| 11 |
Hb_000320_380 |
0.0650320697 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000417_160 |
0.0658097118 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 13 |
Hb_010315_090 |
0.0667896852 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 14 |
Hb_000032_010 |
0.0688528656 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 15 |
Hb_001946_300 |
0.069504942 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 16 |
Hb_007885_070 |
0.0704475506 |
- |
- |
cleavage and polyadenylation specificity factor, putative [Ricinus communis] |
| 17 |
Hb_000120_610 |
0.0714540545 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 18 |
Hb_140049_030 |
0.0740591784 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 19 |
Hb_000264_100 |
0.0747115177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000753_100 |
0.0747632219 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |