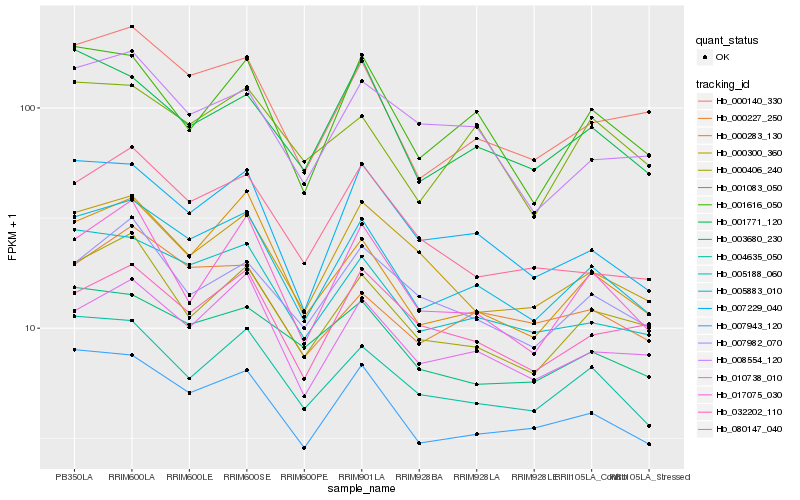
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005883_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007943_120 |
0.0546342981 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 3 |
Hb_005188_060 |
0.0551802009 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 4 |
Hb_000283_130 |
0.0651311893 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 5 |
Hb_010738_010 |
0.0652110058 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 6 |
Hb_000140_330 |
0.0668166041 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 7 |
Hb_007229_040 |
0.0698241924 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 8 |
Hb_004635_050 |
0.0698418203 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 9 |
Hb_000406_240 |
0.0736795853 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_000227_250 |
0.0807042367 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 11 |
Hb_001616_050 |
0.0814593925 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 12 |
Hb_003680_230 |
0.0817268636 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 13 |
Hb_017075_030 |
0.083696927 |
- |
- |
helicase, putative [Ricinus communis] |
| 14 |
Hb_000300_360 |
0.0839398984 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 15 |
Hb_007982_070 |
0.0843822669 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 16 |
Hb_080147_040 |
0.0848702051 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 17 |
Hb_032202_110 |
0.0850490697 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_001771_120 |
0.0870032939 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 19 |
Hb_008554_120 |
0.0883838797 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 20 |
Hb_001083_050 |
0.0888448726 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |