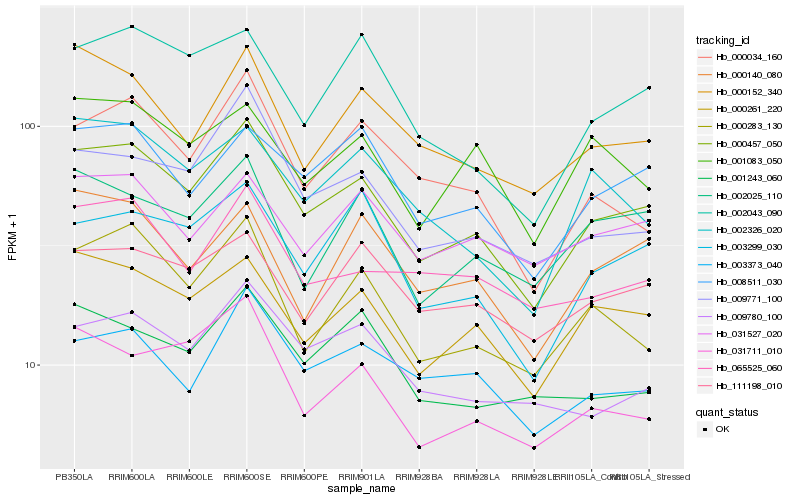
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_050 |
0.0 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 2 |
Hb_000261_220 |
0.0661048711 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 3 |
Hb_008511_030 |
0.0766060815 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 4 |
Hb_003299_030 |
0.080964401 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000283_130 |
0.0813974612 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 6 |
Hb_009771_100 |
0.0817770613 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 7 |
Hb_000152_340 |
0.0869877124 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 8 |
Hb_002043_090 |
0.0882303608 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 9 |
Hb_031711_010 |
0.0882882612 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_009780_100 |
0.0885296659 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 11 |
Hb_003373_040 |
0.0888279209 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 12 |
Hb_002025_110 |
0.0891051261 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_111198_010 |
0.0898743139 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_031527_020 |
0.0913811485 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 15 |
Hb_000140_080 |
0.0914088939 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000034_160 |
0.0914936414 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 17 |
Hb_002326_020 |
0.0915297818 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 18 |
Hb_065525_060 |
0.0917849894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001243_060 |
0.0935105138 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 20 |
Hb_001083_050 |
0.0936009461 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |