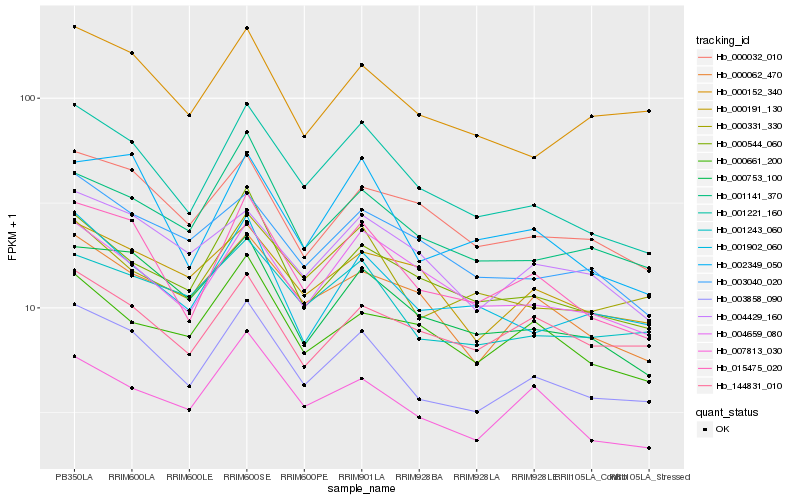
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003858_090 |
0.0 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 2 |
Hb_015475_020 |
0.0609441614 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 3 |
Hb_001221_160 |
0.0663281562 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 4 |
Hb_001243_060 |
0.0731276208 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 5 |
Hb_000753_100 |
0.0802393622 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001141_370 |
0.0808663751 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 7 |
Hb_000152_340 |
0.0815641644 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 8 |
Hb_001902_060 |
0.0833353774 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 9 |
Hb_007813_030 |
0.0849833933 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 10 |
Hb_000544_060 |
0.0855213715 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_000062_470 |
0.0857976478 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 12 |
Hb_002349_050 |
0.0886130714 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 13 |
Hb_000032_010 |
0.0897213186 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 14 |
Hb_004429_160 |
0.0899143519 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000331_330 |
0.0908015543 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000191_130 |
0.0911213044 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 17 |
Hb_144831_010 |
0.0956886015 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004659_080 |
0.0961919343 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 19 |
Hb_003040_020 |
0.0968475346 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 20 |
Hb_000661_200 |
0.0989736618 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |