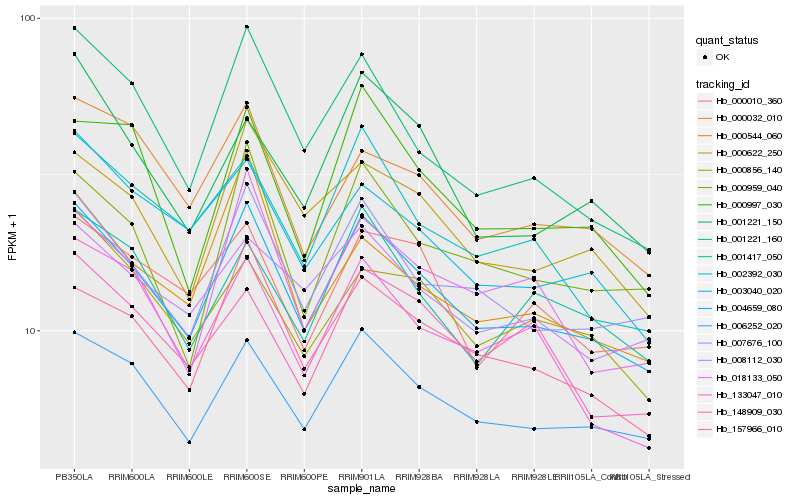
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004659_080 |
0.0 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 2 |
Hb_006252_020 |
0.0522972964 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007676_100 |
0.0523489069 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 4 |
Hb_157966_010 |
0.0555082198 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 5 |
Hb_000622_250 |
0.0645830863 |
- |
- |
atpob1, putative [Ricinus communis] |
| 6 |
Hb_001221_160 |
0.0651991565 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_002392_030 |
0.0701391656 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 8 |
Hb_000032_010 |
0.0706902609 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 9 |
Hb_003040_020 |
0.0713828032 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 10 |
Hb_008112_030 |
0.0734073916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000959_040 |
0.0745353287 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 12 |
Hb_001417_050 |
0.0745752738 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 13 |
Hb_000856_140 |
0.0749340398 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_150 |
0.0764639282 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 15 |
Hb_018133_050 |
0.0769092159 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 16 |
Hb_000997_030 |
0.0770176438 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 17 |
Hb_000544_060 |
0.0770428168 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 18 |
Hb_000010_360 |
0.0773834432 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 19 |
Hb_133047_010 |
0.0778194001 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_148909_030 |
0.0804343588 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |