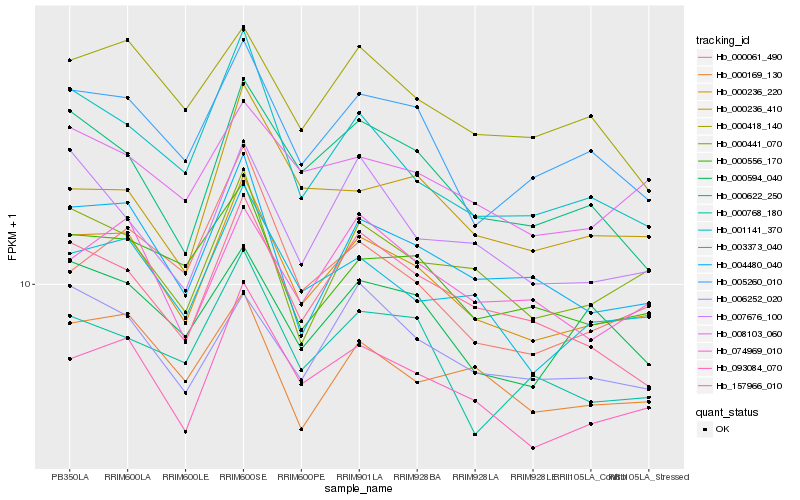
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_220 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 2 |
Hb_093084_070 |
0.0417398127 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 3 |
Hb_003373_040 |
0.0561166203 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 4 |
Hb_004480_040 |
0.0631988423 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005260_010 |
0.0634552197 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001141_370 |
0.0694824642 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 7 |
Hb_000441_070 |
0.0720270159 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 8 |
Hb_157966_010 |
0.0734067798 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 9 |
Hb_000556_170 |
0.0738920332 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000622_250 |
0.0750499316 |
- |
- |
atpob1, putative [Ricinus communis] |
| 11 |
Hb_074969_010 |
0.0778428963 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 12 |
Hb_008103_060 |
0.0780488153 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_000768_180 |
0.0784985187 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 14 |
Hb_000169_130 |
0.0789474824 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 15 |
Hb_000594_040 |
0.0815216025 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 16 |
Hb_000418_140 |
0.0826030534 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 17 |
Hb_000236_410 |
0.0832466082 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 18 |
Hb_007676_100 |
0.0836689964 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 19 |
Hb_000061_490 |
0.0839104637 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 20 |
Hb_006252_020 |
0.0839488131 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |