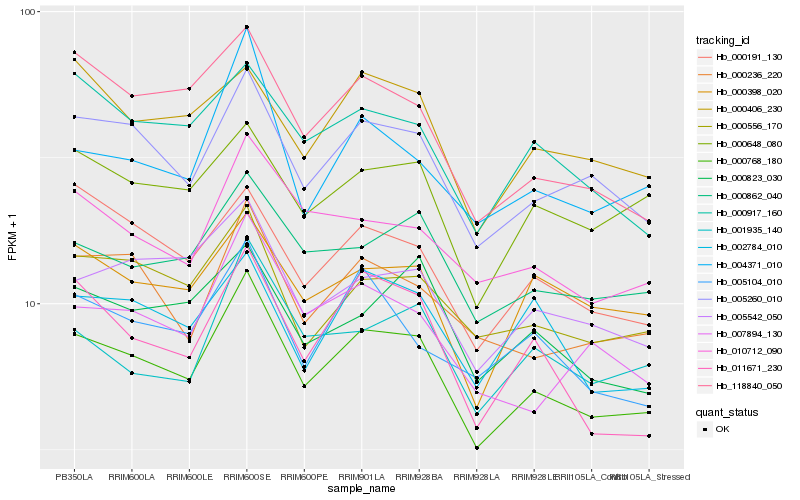
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000768_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 2 |
Hb_005260_010 |
0.0565641351 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000862_040 |
0.0729828321 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_000556_170 |
0.075216287 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005542_050 |
0.0779849074 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 6 |
Hb_000236_220 |
0.0784985187 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 7 |
Hb_010712_090 |
0.0795002815 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 8 |
Hb_000191_130 |
0.0802308354 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 9 |
Hb_000648_080 |
0.0803410848 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 10 |
Hb_118840_050 |
0.0809097285 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 11 |
Hb_000398_020 |
0.0813593226 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004371_010 |
0.0819606399 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 13 |
Hb_000917_160 |
0.0820120018 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001935_140 |
0.0842441111 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 15 |
Hb_000406_230 |
0.0845990815 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 16 |
Hb_011671_230 |
0.0846731839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000823_030 |
0.0849776768 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 18 |
Hb_002784_010 |
0.0853649983 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 19 |
Hb_007894_130 |
0.0858535696 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 20 |
Hb_005104_010 |
0.0861754851 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |