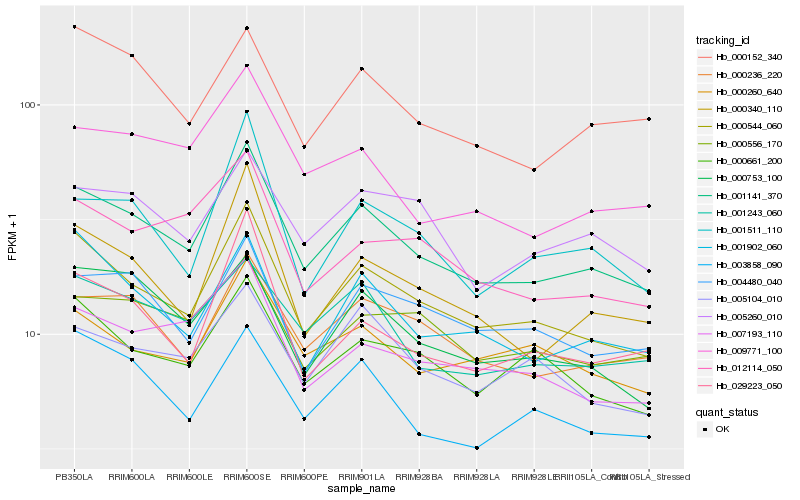
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_370 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 2 |
Hb_000544_060 |
0.046695767 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 3 |
Hb_000236_220 |
0.0694824642 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 4 |
Hb_001243_060 |
0.0740885493 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 5 |
Hb_009771_100 |
0.0773746149 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 6 |
Hb_000340_110 |
0.0785411412 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 7 |
Hb_003858_090 |
0.0808663751 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 8 |
Hb_001902_060 |
0.0815518291 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000753_100 |
0.0816748063 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000152_340 |
0.0826254289 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 11 |
Hb_000661_200 |
0.0827906953 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005260_010 |
0.0828816168 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000260_640 |
0.0834656378 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 14 |
Hb_007193_110 |
0.0843370896 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 15 |
Hb_004480_040 |
0.0846612653 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005104_010 |
0.0856671897 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 17 |
Hb_001511_110 |
0.0860890202 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_029223_050 |
0.0868392712 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000556_170 |
0.0868888065 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 20 |
Hb_012114_050 |
0.0870617394 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |