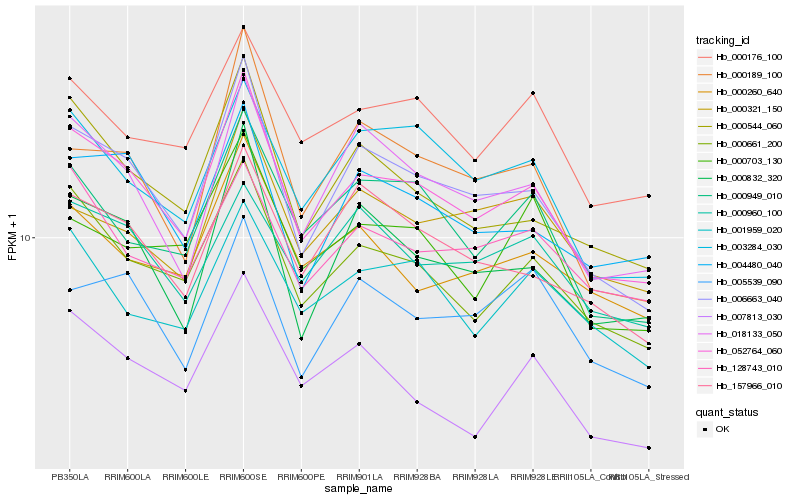
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_060 |
0.0 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_006663_040 |
0.0444042229 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000661_200 |
0.0545341197 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000544_060 |
0.0644689893 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 5 |
Hb_018133_050 |
0.0673054344 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 6 |
Hb_000832_320 |
0.0682104108 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_004480_040 |
0.0733509593 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000949_010 |
0.0751623822 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 9 |
Hb_000176_100 |
0.0758439125 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 10 |
Hb_157966_010 |
0.0762544837 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 11 |
Hb_003284_030 |
0.0762786955 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000260_640 |
0.0788754473 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 13 |
Hb_007813_030 |
0.080600467 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 14 |
Hb_128743_010 |
0.0814530773 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001959_020 |
0.0837110578 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000189_100 |
0.0847374816 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 17 |
Hb_000960_100 |
0.0856636059 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 18 |
Hb_005539_090 |
0.086226161 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000321_150 |
0.0868682079 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000703_130 |
0.0871911778 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |