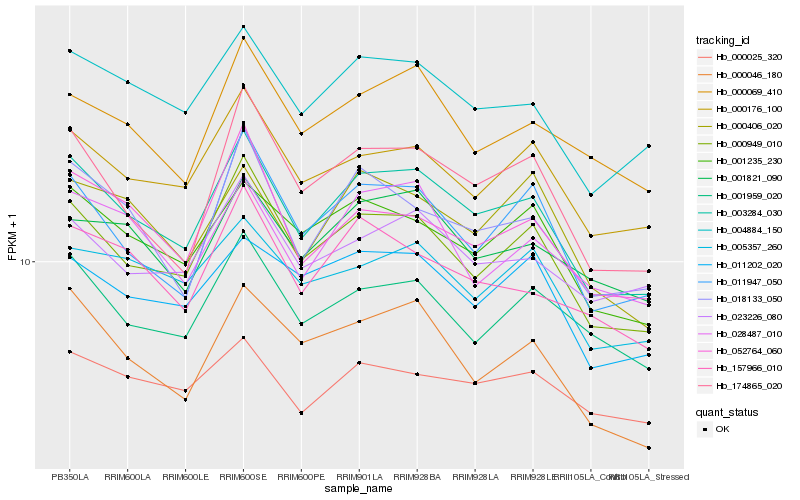
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003284_030 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000949_010 |
0.0492375884 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 3 |
Hb_004884_150 |
0.0605478856 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 4 |
Hb_174865_020 |
0.0669294308 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 5 |
Hb_000176_100 |
0.0674665828 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 6 |
Hb_001235_230 |
0.0685417786 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 7 |
Hb_018133_050 |
0.0705075477 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 8 |
Hb_011947_050 |
0.0713144364 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 9 |
Hb_001959_020 |
0.0736052286 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 10 |
Hb_052764_060 |
0.0762786955 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 11 |
Hb_157966_010 |
0.0767109371 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 12 |
Hb_023226_080 |
0.076978801 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000069_410 |
0.0781629826 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 14 |
Hb_001821_090 |
0.0787633194 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 15 |
Hb_011202_020 |
0.0794121369 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000046_180 |
0.080576033 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 17 |
Hb_005357_260 |
0.0806821323 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 18 |
Hb_000025_320 |
0.080777138 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 19 |
Hb_028487_010 |
0.0811706739 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000406_020 |
0.0820144772 |
- |
- |
- |