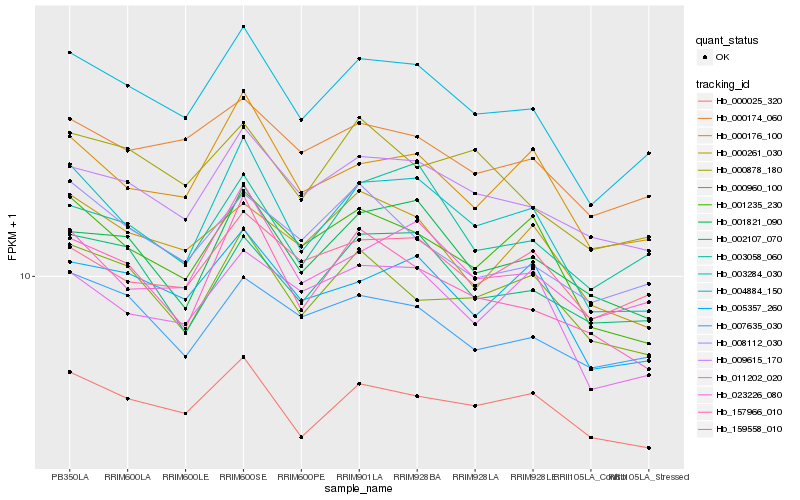
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004884_150 |
0.0 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 2 |
Hb_009615_170 |
0.0594838359 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003284_030 |
0.0605478856 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000174_060 |
0.0651616899 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_005357_260 |
0.0687973848 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 6 |
Hb_000025_320 |
0.0695269586 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 7 |
Hb_023226_080 |
0.0696722211 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000176_100 |
0.0711651182 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 9 |
Hb_007635_030 |
0.0719106213 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_001821_090 |
0.0740379778 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 11 |
Hb_000878_180 |
0.0743282804 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008112_030 |
0.075167467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003058_060 |
0.0751965741 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 14 |
Hb_002107_070 |
0.0760788134 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 15 |
Hb_001235_230 |
0.0766953809 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 16 |
Hb_159558_010 |
0.0770883523 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 17 |
Hb_157966_010 |
0.0771204743 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 18 |
Hb_011202_020 |
0.0783837709 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000960_100 |
0.0785718259 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 20 |
Hb_000261_030 |
0.0791018144 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |