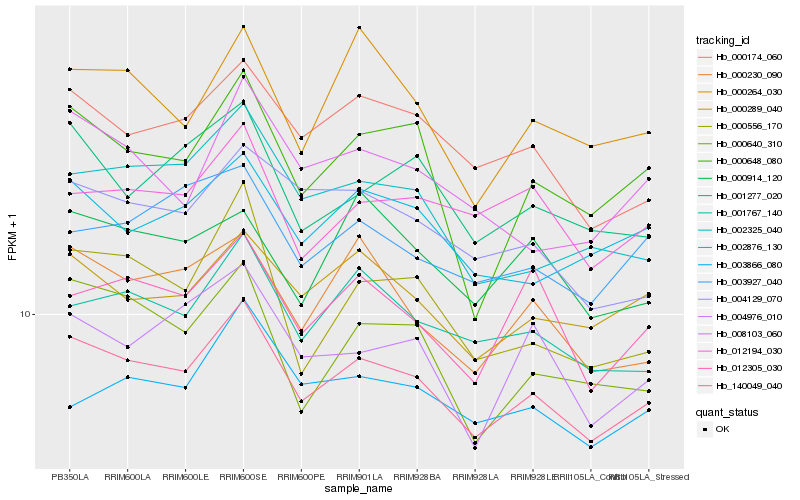
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140049_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 2 |
Hb_000174_060 |
0.0495738648 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_000289_040 |
0.0510996601 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 4 |
Hb_001767_140 |
0.0531974985 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 5 |
Hb_002876_130 |
0.0559086767 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 6 |
Hb_000230_090 |
0.0588479821 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001277_020 |
0.0593794042 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 8 |
Hb_012305_030 |
0.0611212973 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 9 |
Hb_004129_070 |
0.0613629454 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 10 |
Hb_003927_040 |
0.0618111666 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 11 |
Hb_002325_040 |
0.0618952946 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000640_310 |
0.0622557526 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 13 |
Hb_012194_030 |
0.0624907046 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_004976_010 |
0.0635118215 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 15 |
Hb_003866_080 |
0.0637516789 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 16 |
Hb_000648_080 |
0.0638954792 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_000264_030 |
0.0640668462 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 18 |
Hb_008103_060 |
0.0641996277 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_000556_170 |
0.0646379193 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000914_120 |
0.0648129022 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |