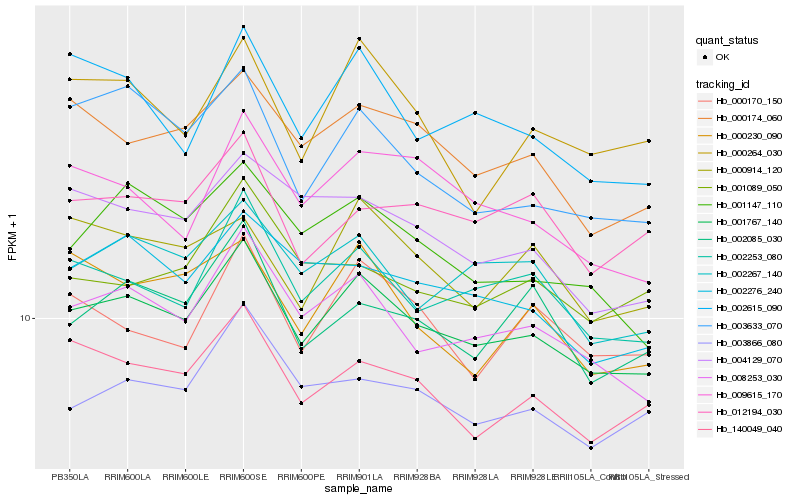
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001767_140 |
0.0 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 2 |
Hb_008253_030 |
0.0406992877 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 3 |
Hb_002253_080 |
0.0466438063 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 4 |
Hb_002267_140 |
0.0527616533 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 5 |
Hb_140049_040 |
0.0531974985 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 6 |
Hb_000174_060 |
0.0535393605 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_003866_080 |
0.055278044 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 8 |
Hb_002276_240 |
0.0573741659 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 9 |
Hb_002615_090 |
0.0600981284 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001147_110 |
0.0605042247 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 11 |
Hb_002085_030 |
0.0605233672 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004129_070 |
0.0609025764 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 13 |
Hb_009615_170 |
0.0610323901 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000170_150 |
0.0612058792 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 15 |
Hb_012194_030 |
0.0622536725 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000914_120 |
0.0629475612 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003633_070 |
0.0647216802 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 18 |
Hb_000230_090 |
0.0655310549 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001089_050 |
0.0660501987 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_000264_030 |
0.0663721814 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |