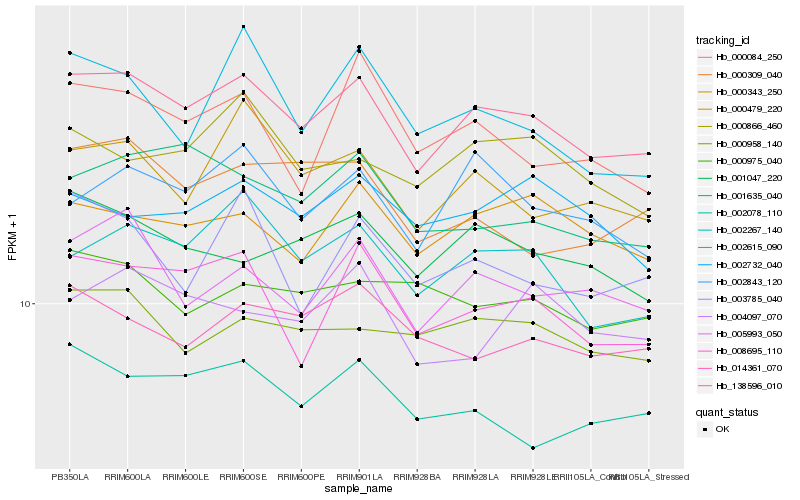
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138596_010 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 2 |
Hb_002267_140 |
0.0555505684 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 3 |
Hb_008695_110 |
0.0581061787 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005993_050 |
0.0586721271 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 5 |
Hb_000479_220 |
0.0603216378 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 6 |
Hb_002843_120 |
0.0605859609 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000343_250 |
0.0622720827 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 8 |
Hb_001047_220 |
0.0632028743 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 9 |
Hb_002078_110 |
0.0639793061 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 10 |
Hb_000958_140 |
0.0640459506 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 11 |
Hb_001635_040 |
0.0654263702 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 12 |
Hb_000866_460 |
0.0675026556 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_014361_070 |
0.0682821527 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 14 |
Hb_000084_250 |
0.0695920817 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_000309_040 |
0.070504925 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 16 |
Hb_000975_040 |
0.0717394549 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 17 |
Hb_003785_040 |
0.072117329 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004097_070 |
0.0724340298 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 19 |
Hb_002615_090 |
0.0727598454 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002732_040 |
0.0731498634 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |