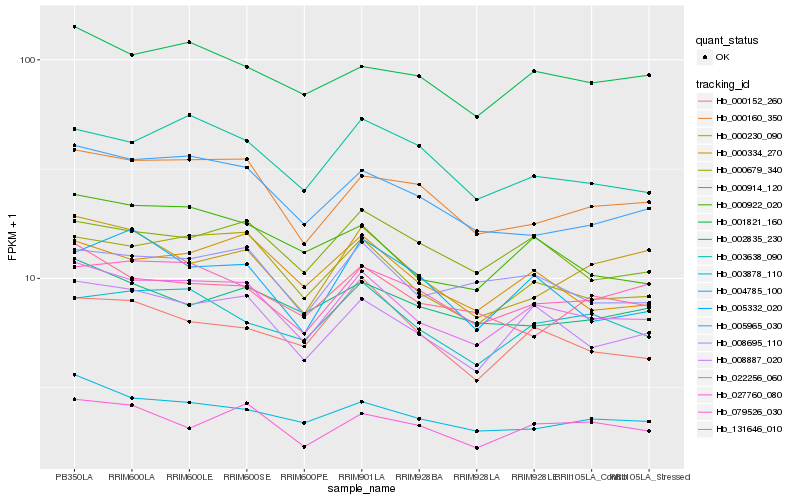
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022256_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 2 |
Hb_008887_020 |
0.0443181312 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 3 |
Hb_000679_340 |
0.0490797507 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008695_110 |
0.0617931972 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000230_090 |
0.0646111637 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000160_350 |
0.0669186448 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 7 |
Hb_001821_160 |
0.0671428136 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 8 |
Hb_000914_120 |
0.0674851536 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 9 |
Hb_079526_030 |
0.0677168711 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 10 |
Hb_027760_080 |
0.0682634823 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_005965_030 |
0.06833026 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 12 |
Hb_003878_110 |
0.068562853 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 13 |
Hb_004785_100 |
0.0687116407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000922_020 |
0.0704188741 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003638_090 |
0.070991282 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 16 |
Hb_000152_260 |
0.0743802009 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 17 |
Hb_002835_230 |
0.0781653189 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000334_270 |
0.0792542986 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_131646_010 |
0.0804793707 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005332_020 |
0.0807399394 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |