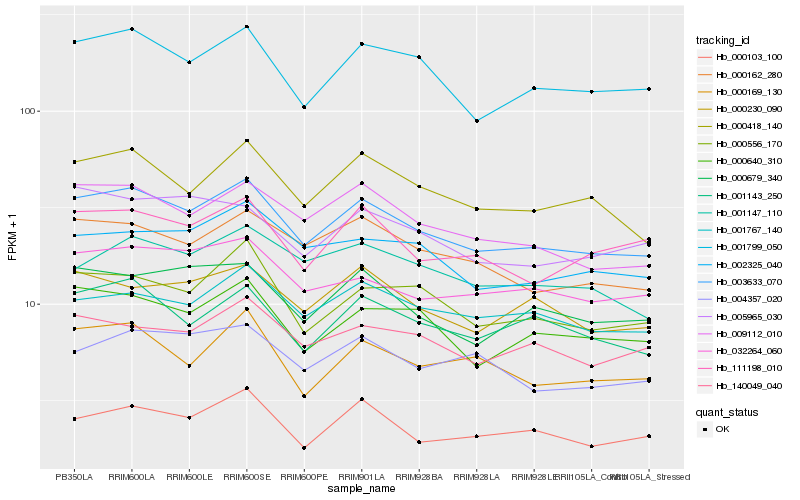
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003633_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 2 |
Hb_001799_050 |
0.0511589373 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 3 |
Hb_000418_140 |
0.0538189214 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 4 |
Hb_009112_010 |
0.0549359492 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 5 |
Hb_000162_280 |
0.0602896163 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 6 |
Hb_001143_250 |
0.0610488692 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 7 |
Hb_000640_310 |
0.061689125 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 8 |
Hb_000556_170 |
0.0625572807 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 9 |
Hb_032264_060 |
0.0629549713 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 10 |
Hb_000679_340 |
0.0644954762 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001767_140 |
0.0647216802 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 12 |
Hb_140049_040 |
0.0653455638 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 13 |
Hb_000230_090 |
0.0656775128 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002325_040 |
0.0663441555 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 15 |
Hb_111198_010 |
0.0675755001 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000103_100 |
0.0689977643 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000169_130 |
0.0706725343 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 18 |
Hb_004357_020 |
0.0706766066 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 19 |
Hb_001147_110 |
0.0709437665 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 20 |
Hb_005965_030 |
0.0734425135 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |