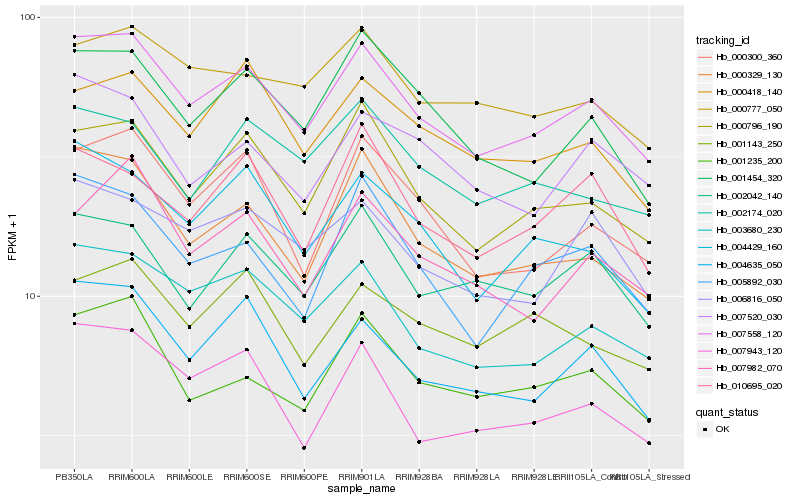
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007558_120 |
0.0 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 2 |
Hb_000796_190 |
0.0509107681 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 3 |
Hb_003680_230 |
0.0542507896 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 4 |
Hb_000329_130 |
0.0604291395 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 5 |
Hb_006816_050 |
0.0626349002 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 6 |
Hb_004635_050 |
0.0631785466 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 7 |
Hb_002042_140 |
0.0653986416 |
- |
- |
PREDICTED: uncharacterized protein LOC105647570 [Jatropha curcas] |
| 8 |
Hb_001454_320 |
0.0661510379 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000300_360 |
0.0672135952 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 10 |
Hb_000777_050 |
0.0684625042 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 11 |
Hb_001235_200 |
0.068524567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000418_140 |
0.0687151243 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 13 |
Hb_004429_160 |
0.0688665609 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007982_070 |
0.070177875 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 15 |
Hb_005892_030 |
0.0707425826 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 16 |
Hb_007943_120 |
0.0717776729 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 17 |
Hb_007520_030 |
0.0727150505 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 18 |
Hb_002174_020 |
0.0730310265 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_001143_250 |
0.0734112144 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 20 |
Hb_010695_020 |
0.0740413393 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 [Jatropha curcas] |