| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
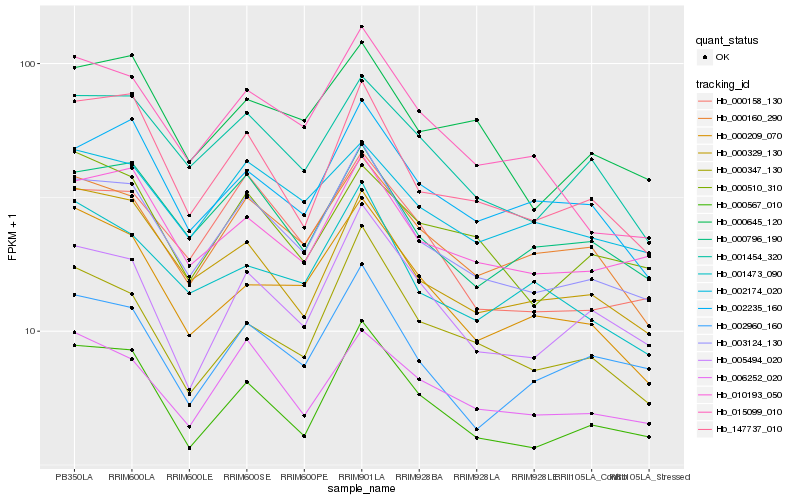
Hb_003124_130 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000567_010 |
0.0480031587 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 3 |
Hb_000796_190 |
0.0601665974 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 4 |
Hb_147737_010 |
0.0602987753 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001454_320 |
0.0614592512 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000158_130 |
0.0621751015 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000160_290 |
0.0625433605 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_010193_050 |
0.0628776668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000645_120 |
0.0674211306 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 10 |
Hb_002174_020 |
0.0690867496 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 11 |
Hb_005494_020 |
0.0695141557 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 12 |
Hb_000329_130 |
0.070137995 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 13 |
Hb_006252_020 |
0.070144152 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000347_130 |
0.0715353435 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 15 |
Hb_002235_160 |
0.0719012539 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 16 |
Hb_015099_010 |
0.0721139201 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 17 |
Hb_000510_310 |
0.0747440434 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 18 |
Hb_001473_090 |
0.0750910284 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 19 |
Hb_000209_070 |
0.0762548434 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 20 |
Hb_002960_160 |
0.0762865722 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |