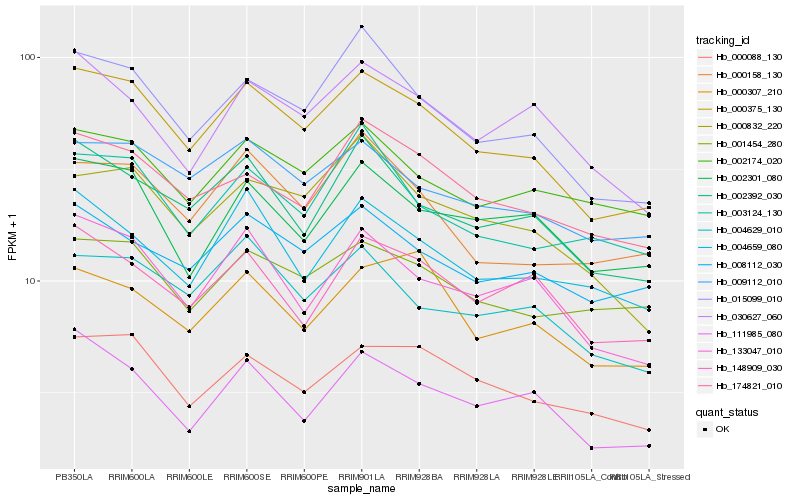
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000375_130 |
0.0 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_015099_010 |
0.0593904482 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 3 |
Hb_000088_130 |
0.0707549342 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 4 |
Hb_002392_030 |
0.0711699677 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 5 |
Hb_133047_010 |
0.0720312376 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_009112_010 |
0.0729945325 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 7 |
Hb_174821_010 |
0.0766791394 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 8 |
Hb_004629_010 |
0.0771376834 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 9 |
Hb_000832_220 |
0.0798391965 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000158_130 |
0.0799356431 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008112_030 |
0.0819262762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002301_080 |
0.0837364201 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004659_080 |
0.0863096059 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 14 |
Hb_148909_030 |
0.0866036974 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001454_280 |
0.0869606646 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 16 |
Hb_111985_080 |
0.0872717152 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 17 |
Hb_002174_020 |
0.0877164396 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_003124_130 |
0.0881111871 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 19 |
Hb_030627_060 |
0.0887365822 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 20 |
Hb_000307_210 |
0.0890810682 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |