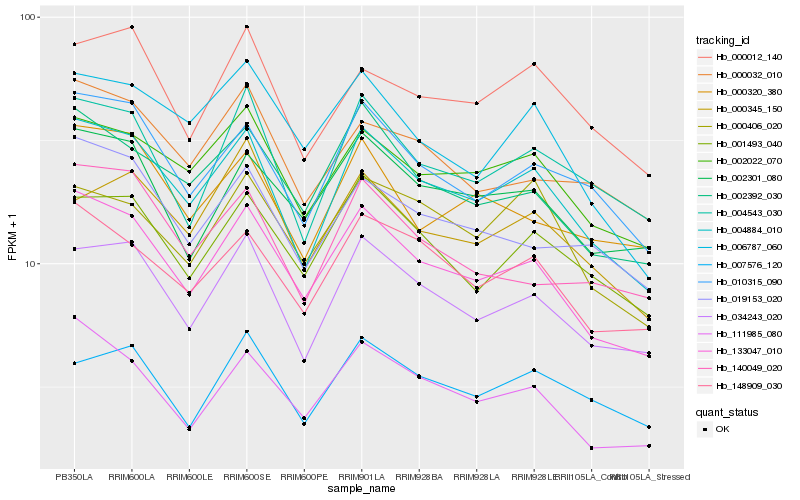
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004884_010 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 2 |
Hb_133047_010 |
0.0546383231 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_034243_020 |
0.0571789236 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010315_090 |
0.0624133074 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 5 |
Hb_002022_070 |
0.0642885015 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 6 |
Hb_148909_030 |
0.0723481469 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004543_030 |
0.0757506589 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 8 |
Hb_002392_030 |
0.0783313788 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 9 |
Hb_001493_040 |
0.0793506272 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 10 |
Hb_000012_140 |
0.0802230629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_019153_020 |
0.0836621003 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 12 |
Hb_111985_080 |
0.0843152255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 13 |
Hb_006787_060 |
0.0883105905 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000406_020 |
0.089135491 |
- |
- |
- |
| 15 |
Hb_000032_010 |
0.0893867085 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 16 |
Hb_002301_080 |
0.0902205692 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000345_150 |
0.0903336825 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000320_380 |
0.0903521489 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 19 |
Hb_140049_020 |
0.092269958 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 20 |
Hb_007576_120 |
0.0923888464 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |