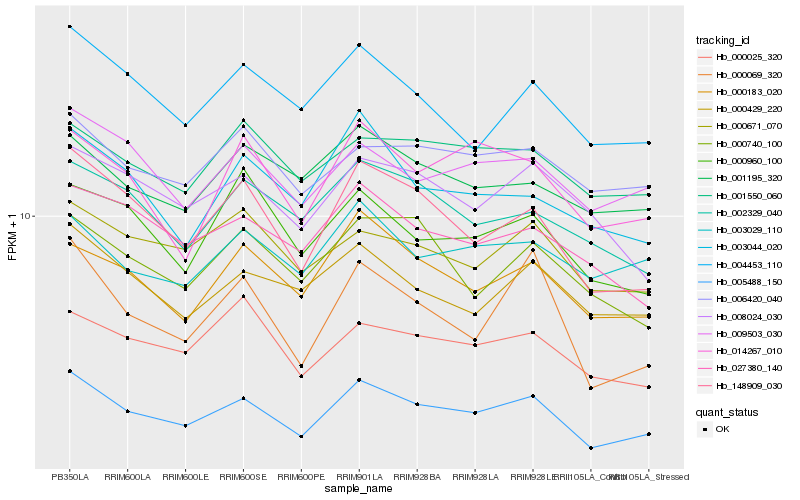
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005488_150 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_148909_030 |
0.0569662974 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000183_020 |
0.0759521459 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000069_320 |
0.0771849514 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 5 |
Hb_004453_110 |
0.0796904617 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 6 |
Hb_009503_030 |
0.0798247726 |
- |
- |
protein dimerization, putative [Ricinus communis] |
| 7 |
Hb_000429_220 |
0.0802245565 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000671_070 |
0.0818905509 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 9 |
Hb_000025_320 |
0.0828024948 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 10 |
Hb_027380_140 |
0.0834245498 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 11 |
Hb_014267_010 |
0.0841230813 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002329_040 |
0.0855043372 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 13 |
Hb_008024_030 |
0.0857978007 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 14 |
Hb_006420_040 |
0.0858381428 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003044_020 |
0.0862992679 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 16 |
Hb_000740_100 |
0.0871580077 |
- |
- |
calpain, putative [Ricinus communis] |
| 17 |
Hb_000960_100 |
0.0875300834 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 18 |
Hb_001550_060 |
0.0882672315 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_003029_110 |
0.0891015528 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001195_320 |
0.0893337791 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |