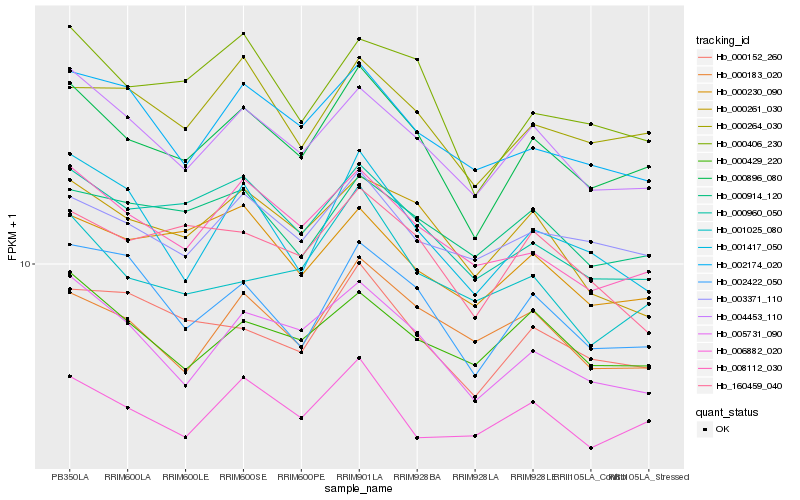
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000896_080 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP95 isoform X1 [Vitis vinifera] |
| 2 |
Hb_004453_110 |
0.0598400767 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 3 |
Hb_005731_090 |
0.0685770797 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 4 |
Hb_000406_230 |
0.07073113 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 5 |
Hb_000264_030 |
0.0708129926 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 6 |
Hb_008112_030 |
0.0712234479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000960_050 |
0.0719822829 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 8 |
Hb_002422_050 |
0.0750447656 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000152_260 |
0.0753638918 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 10 |
Hb_003371_110 |
0.076384747 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 11 |
Hb_006882_020 |
0.0766567707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002174_020 |
0.0792853274 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_000429_220 |
0.0799772425 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000914_120 |
0.0817705373 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001417_050 |
0.0826851825 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 16 |
Hb_000230_090 |
0.0829269338 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000261_030 |
0.0832208783 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 18 |
Hb_000183_020 |
0.0846416994 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 19 |
Hb_160459_040 |
0.0856580303 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 20 |
Hb_001025_080 |
0.0858306865 |
- |
- |
hypothetical protein JCGZ_11431 [Jatropha curcas] |