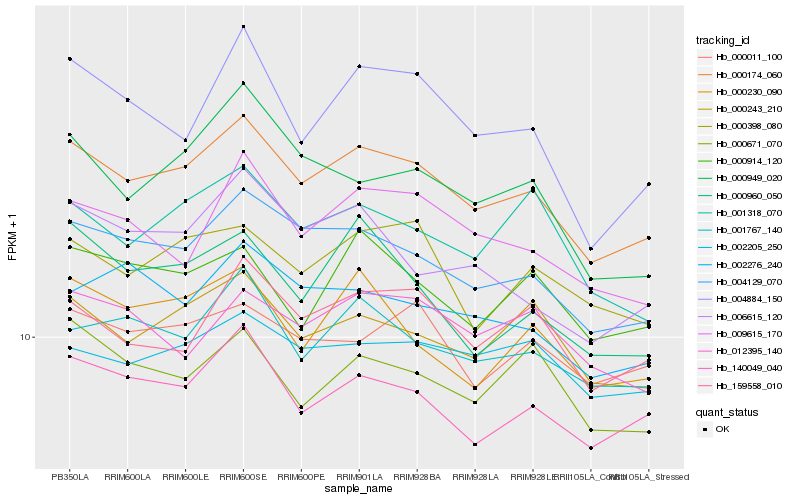
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_060 |
0.0 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 2 |
Hb_004129_070 |
0.0389276753 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 3 |
Hb_140049_040 |
0.0495738648 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 4 |
Hb_000243_210 |
0.0518726676 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_001767_140 |
0.0535393605 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 6 |
Hb_159558_010 |
0.0571878454 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 7 |
Hb_009615_170 |
0.0603720097 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000398_080 |
0.0605706099 |
- |
- |
tip120, putative [Ricinus communis] |
| 9 |
Hb_002205_250 |
0.061327576 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000230_090 |
0.0618370275 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002276_240 |
0.0622156671 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 12 |
Hb_001318_070 |
0.0625387197 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000011_100 |
0.063067414 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 14 |
Hb_000671_070 |
0.0637136156 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 15 |
Hb_000914_120 |
0.0639052982 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004884_150 |
0.0651616899 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 17 |
Hb_006615_120 |
0.0657119167 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 18 |
Hb_000960_050 |
0.0662240316 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 19 |
Hb_000949_020 |
0.0667205132 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 20 |
Hb_012395_140 |
0.0669955392 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |