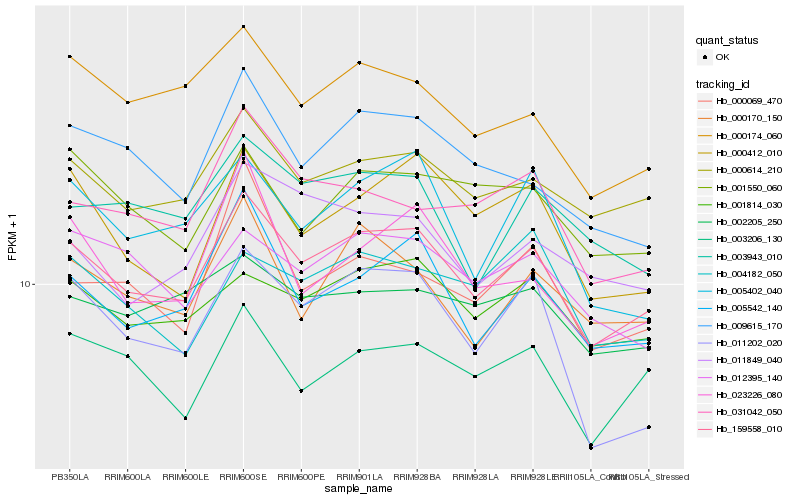
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159558_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 2 |
Hb_000614_210 |
0.0470527786 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005542_140 |
0.0509980571 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 4 |
Hb_001814_030 |
0.0526037068 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 5 |
Hb_000174_060 |
0.0571878454 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_003206_130 |
0.0584472644 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 7 |
Hb_009615_170 |
0.0596260678 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000170_150 |
0.0597227486 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000412_010 |
0.0603024119 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_031042_050 |
0.0609713469 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 11 |
Hb_012395_140 |
0.0615376056 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 12 |
Hb_002205_250 |
0.0623479339 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001550_060 |
0.0627802756 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 14 |
Hb_005402_040 |
0.0628283636 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 15 |
Hb_011849_040 |
0.0636290393 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_003943_010 |
0.0637879275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023226_080 |
0.0649870761 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004182_050 |
0.0662502872 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011202_020 |
0.0667252996 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000069_470 |
0.0667546233 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |