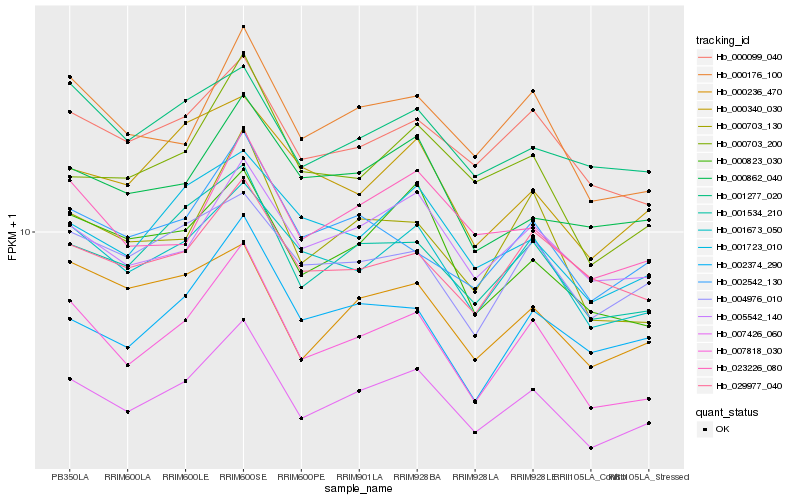
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_007818_030 |
0.0502435002 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 3 |
Hb_001534_210 |
0.0507499548 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001673_050 |
0.0582012965 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005542_140 |
0.0587571999 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 6 |
Hb_000099_040 |
0.0634851419 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000176_100 |
0.0656613291 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 8 |
Hb_000703_200 |
0.0671656796 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001723_010 |
0.0675782967 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 10 |
Hb_023226_080 |
0.0690026037 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002542_130 |
0.070569562 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 12 |
Hb_000823_030 |
0.0719525853 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 13 |
Hb_004976_010 |
0.072112143 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 14 |
Hb_000862_040 |
0.0726382618 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_000703_130 |
0.073183079 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 16 |
Hb_000340_030 |
0.0744571807 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 17 |
Hb_000236_470 |
0.0747049259 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 18 |
Hb_002374_290 |
0.0750217561 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 19 |
Hb_029977_040 |
0.075762686 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 20 |
Hb_001277_020 |
0.0766431846 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |