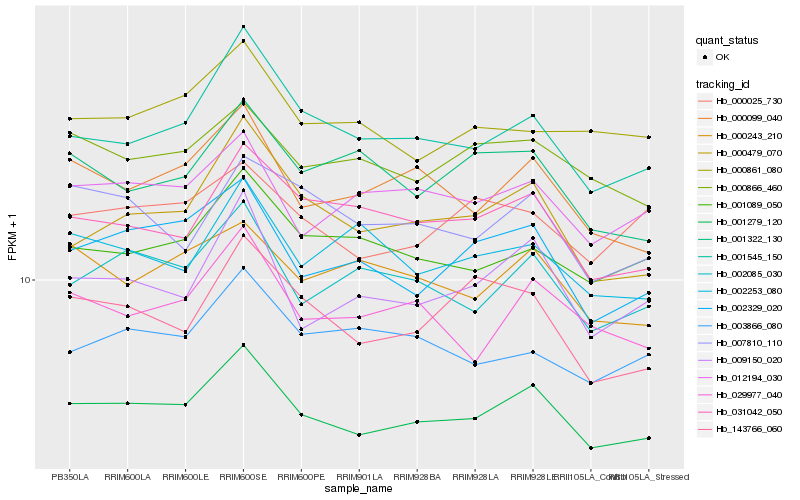
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_120 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 2 |
Hb_001545_150 |
0.057127796 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 3 |
Hb_000099_040 |
0.0583353732 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_002329_020 |
0.0584210352 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 5 |
Hb_009150_020 |
0.0607255628 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 6 |
Hb_002085_030 |
0.0607389968 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012194_030 |
0.0620223048 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000479_070 |
0.0627133612 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 9 |
Hb_000866_460 |
0.0633058956 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_031042_050 |
0.0636261959 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 11 |
Hb_029977_040 |
0.0639302599 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 12 |
Hb_001089_050 |
0.0660768255 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_000243_210 |
0.0706061594 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000025_730 |
0.0710349547 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_143766_060 |
0.0719666591 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 16 |
Hb_007810_110 |
0.072106796 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 17 |
Hb_000861_080 |
0.0732622395 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 18 |
Hb_002253_080 |
0.07591418 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 19 |
Hb_001322_130 |
0.0760480271 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 20 |
Hb_003866_080 |
0.0762338361 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |